Genome-enabled prediction of genetic values using radial basis function neural networks
- PMID: 22566067
- PMCID: PMC3405257
- DOI: 10.1007/s00122-012-1868-9
Genome-enabled prediction of genetic values using radial basis function neural networks
Abstract
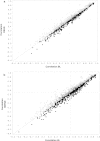
The availability of high density panels of molecular markers has prompted the adoption of genomic selection (GS) methods in animal and plant breeding. In GS, parametric, semi-parametric and non-parametric regressions models are used for predicting quantitative traits. This article shows how to use neural networks with radial basis functions (RBFs) for prediction with dense molecular markers. We illustrate the use of the linear Bayesian LASSO regression model and of two non-linear regression models, reproducing kernel Hilbert spaces (RKHS) regression and radial basis function neural networks (RBFNN) on simulated data and real maize lines genotyped with 55,000 markers and evaluated for several trait-environment combinations. The empirical results of this study indicated that the three models showed similar overall prediction accuracy, with a slight and consistent superiority of RKHS and RBFNN over the additive Bayesian LASSO model. Results from the simulated data indicate that RKHS and RBFNN models captured epistatic effects; however, adding non-signal (redundant) predictors (interaction between markers) can adversely affect the predictive accuracy of the non-linear regression models.
Figures



References
-
- Bernardo R. Breeding for quantitative traits in plants. Minneapolis: Stemma Press; 2002.
-
- Broomhead DS, Lowe D. Multivariable functional interpolation and adaptive networks. Complex Syst. 1988;2:321–355.
-
- Crossa J, de los Campos G, Pérez P, Gianola D, Burgueño J, Araus JL, Makumbi D, Dreisigacker S, Yan J, Arief V, Banziger M, Braun H-J. Prediction of genetic values of quantitative traits in plant breeding using pedigree and molecular markers. Genetics. 2010;186:713–724. doi: 10.1534/genetics.110.118521. - DOI - PMC - PubMed
-
- Crossa J, Pérez P, de los Campos G, Mahuku G, Dreisigacker S, Magorokosho C. Genomic selection and prediction in plant breeding. J Crop Improv. 2011;25(3):239–261. doi: 10.1080/15427528.2011.558767. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources

