RIP1, a member of an Arabidopsis protein family, interacts with the protein RARE1 and broadly affects RNA editing
- PMID: 22566615
- PMCID: PMC3365174
- DOI: 10.1073/pnas.1121465109
RIP1, a member of an Arabidopsis protein family, interacts with the protein RARE1 and broadly affects RNA editing
Abstract
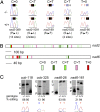
Transcripts of plant organelle genes are modified by cytidine-to-uridine (C-to-U) RNA editing, often changing the encoded amino acid predicted from the DNA sequence. Members of the PLS subclass of the pentatricopeptide repeat (PPR) motif-containing family are site-specific recognition factors for either chloroplast or mitochondrial C targets of editing. However, other than PPR proteins and the cis-elements on the organelle transcripts, no other components of the editing machinery in either organelle have previously been identified. The Arabidopsis chloroplast PPR protein Required for AccD RNA Editing 1 (RARE1) specifies editing of a C in the accD transcript. RARE1 was detected in a complex of >200 kDa. We immunoprecipitated epitope-tagged RARE1, and tandem MS/MS analysis identified a protein of unknown function lacking PPR motifs; we named it RNA-editing factor interacting protein 1 (RIP1). Yeast two-hybrid analysis confirmed RIP1 interaction with RARE1, and RIP1-GFP fusions were found in both chloroplasts and mitochondria. Editing assays for all 34 known Arabidopsis chloroplast targets in a rip1 mutant revealed altered efficiency of 14 editing events. In mitochondria, 266 editing events were found to have reduced efficiency, with major loss of editing at 108 C targets. Virus-induced gene silencing of RIP1 confirmed the altered editing efficiency. Transient introduction of a WT RIP1 allele into rip1 improved the defective RNA editing. The presence of RIP1 in a protein complex along with chloroplast editing factor RARE1 indicates that RIP1 is an important component of the RNA editing apparatus that acts on many chloroplast and mitochondrial C targets.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Stern DB, Goldschmidt-Clermont M, Hanson MR. Chloroplast RNA metabolism. Annu Rev Plant Biol. 2010;61:125–155. - PubMed
-
- Fujii S, Small I. The evolution of RNA editing and pentatricopeptide repeat genes. New Phytol. 2011;191:37–47. - PubMed
-
- Chateigner-Boutin AL, Small I. Plant RNA editing. RNA Biol. 2010;7:213–219. - PubMed
-
- Covello PS, Gray MW. RNA editing in plant mitochondria. Nature. 1989;341:662–666. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

