A cluster of cooperating tumor-suppressor gene candidates in chromosomal deletions
- PMID: 22566646
- PMCID: PMC3361457
- DOI: 10.1073/pnas.1206062109
A cluster of cooperating tumor-suppressor gene candidates in chromosomal deletions
Abstract
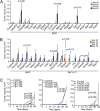
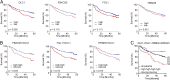
The large chromosomal deletions frequently observed in cancer genomes are often thought to arise as a "two-hit" mechanism in the process of tumor-suppressor gene (TSG) inactivation. Using a murine model system of hepatocellular carcinoma (HCC) and in vivo RNAi, we test an alternative hypothesis, that such deletions can arise from selective pressure to attenuate the activity of multiple genes. By targeting the mouse orthologs of genes frequently deleted on human 8p22 and adjacent regions, which are lost in approximately half of several other major epithelial cancers, we provide evidence suggesting that multiple genes on chromosome 8p can cooperatively inhibit tumorigenesis in mice, and that their cosuppression can synergistically promote tumor growth. In addition, in human HCC patients, the combined down-regulation of functionally validated 8p TSGs is associated with poor survival, in contrast to the down-regulation of any individual gene. Our data imply that large cancer-associated deletions can produce phenotypes distinct from those arising through loss of a single TSG, and as such should be considered and studied as distinct mutational events.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Comment in
-
Significance of clustered tumor suppressor genes in cancer.Future Oncol. 2012 Sep;8(9):1091-3. doi: 10.2217/fon.12.109. Future Oncol. 2012. PMID: 23030484
References
-
- Baker SJ, et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science. 1989;244:217–221. - PubMed
-
- Birnbaum D, et al. Chromosome arm 8p and cancer: A fragile hypothesis. Lancet Oncol. 2003;4:639–642. - PubMed
-
- El Gammal AT, et al. Chromosome 8p deletions and 8q gains are associated with tumor progression and poor prognosis in prostate cancer. Clin Cancer Res. 2010;16:56–64. - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

