Hydroxylation of 5-methylcytosine by TET2 maintains the active state of the mammalian HOXA cluster
- PMID: 22569366
- PMCID: PMC3576573
- DOI: 10.1038/ncomms1826
Hydroxylation of 5-methylcytosine by TET2 maintains the active state of the mammalian HOXA cluster
Abstract
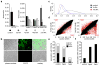
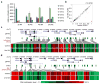
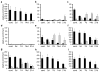
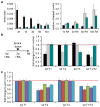
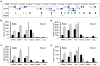
Differentiation is accompanied by extensive epigenomic reprogramming, leading to the repression of stemness factors and the transcriptional maintenance of activated lineage-specific genes. Here we use the mammalian Hoxa cluster of developmental genes as a model system to follow changes in DNA modification patterns during retinoic acid-induced differentiation. We find the inactive cluster to be marked by defined patterns of 5-methylcytosine (5mC). Upon the induction of differentiation, the active anterior part of the cluster becomes increasingly enriched in 5-hydroxymethylcytosine (5hmC), following closely the colinear activation pattern of the gene array, which is paralleled by the reduction of 5mC. Depletion of the 5hmC generating dioxygenase Tet2 impairs the maintenance of Hoxa activity and partially restores 5mC levels. Our results indicate that gene-specific 5mC-5hmC conversion by Tet2 is crucial for the maintenance of active chromatin states at lineage-specific loci.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

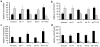

References
-
- Duboule D, Morata G. Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet. 1994;10:358–364. - PubMed
-
- Kmita M, Duboule D. Organizing axes in time and space; 25 years of colinear tinkering. Science. 2003;301:331–333. - PubMed
-
- Soshnikova N, Duboule D. Epigenetic regulation of Hox gene activation: the waltz of methyls. Bioessays. 2008;30:199–202. - PubMed
-
- Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

