Targeted deletion of microRNA-22 promotes stress-induced cardiac dilation and contractile dysfunction
- PMID: 22570371
- PMCID: PMC3503489
- DOI: 10.1161/CIRCULATIONAHA.111.044354
Targeted deletion of microRNA-22 promotes stress-induced cardiac dilation and contractile dysfunction
Abstract
Background: Delineating the role of microRNAs (miRNAs) in the posttranscriptional gene regulation offers new insights into how the heart adapts to pathological stress. We developed a knockout of miR-22 in mice and investigated its function in the heart.
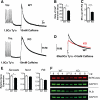
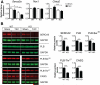
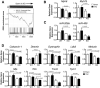
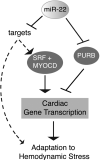
Methods and results: Here, we show that miR-22-deficient mice are impaired in inotropic and lusitropic response to acute stress by dobutamine. Furthermore, the absence of miR-22 sensitized mice to cardiac decompensation and left ventricular dilation after long-term stimulation by pressure overload. Calcium transient analysis revealed reduced sarcoplasmic reticulum Ca(2+) load in association with repressed sarcoplasmic reticulum Ca(2+) ATPase activity in mutant myocytes. Genetic ablation of miR-22 also led to a decrease in cardiac expression levels for Serca2a and muscle-restricted genes encoding proteins in the vicinity of the cardiac Z disk/titin cytoskeleton. These phenotypes were attributed in part to inappropriate repression of serum response factor activity in stressed hearts. Global analysis revealed increased expression of the transcriptional/translational repressor purine-rich element binding protein B, a highly conserved miR-22 target implicated in the negative control of muscle expression.
Conclusion: These data indicate that miR-22 functions as an integrator of Ca(2+) homeostasis and myofibrillar protein content during stress in the heart and shed light on the mechanisms that enhance propensity toward heart failure.
Figures



Comment in
-
Resolving a catch-22 in cardiac gene regulation.Circulation. 2012 Jun 5;125(22):2695-7. doi: 10.1161/CIRCULATIONAHA.112.110767. Epub 2012 May 8. Circulation. 2012. PMID: 22570372 No abstract available.
References
-
- Diwan A, Dorn GW., 2nd Decompensation of cardiac hypertrophy: cellular mechanisms and novel therapeutic targets. Physiology (Bethesda) 2007;22:56–64. - PubMed
-
- Frey N, Olson EN. Cardiac hypertrophy: the good, the bad, and the ugly. Annu Rev Physiol. 2003;65:45–79. - PubMed
-
- Periasamy M, Huke S. SERCA pump level is a critical determinant of Ca(2+)homeostasis and cardiac contractility. J Mol Cell Cardiol. 2001;33:1053–1063. - PubMed
-
- Ahmad F, Seidman JG, Seidman CE. The genetic basis for cardiac remodeling. Annu Rev Genomics Hum Genet. 2005;6:185–216. - PubMed
-
- Bers DM. Cardiac excitation-contraction coupling. Nature. 2002;415:198–205. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

