Signaling in cell differentiation and morphogenesis
- PMID: 22570373
- PMCID: PMC3367549
- DOI: 10.1101/cshperspect.a008151
Signaling in cell differentiation and morphogenesis
Abstract
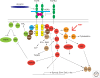
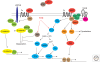
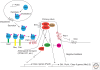
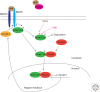
All the information to make a complete, fully functional living organism is encoded within the genome of the fertilized oocyte. How is this genetic code translated into the vast array of cellular behaviors that unfold during the course of embryonic development, as the zygote slowly morphs into a new organism? Studies over the last 30 years or so have shown that many of these cellular processes are driven by secreted or membrane-bound signaling molecules. Elucidating how the genetic code is translated into instructions or signals during embryogenesis, how signals are generated at the correct time and place and at the appropriate level, and finally, how these instructions are interpreted and put into action, are some of the central questions of developmental biology. Our understanding of the causes of congenital malformations and disease has improved substantially with the rapid advances in our knowledge of signaling pathways and their regulation during development. In this article, I review some of the signaling pathways that play essential roles during embryonic development. These examples show some of the mechanisms used by cells to receive and interpret developmental signals. I also discuss how signaling pathways downstream from these signals are regulated and how they induce specific cellular responses that ultimately affect cell fate and morphogenesis.
Figures
References
-
- Airaksinen MS, Holm L, Hatinen T 2006. Evolution of the GDNF family ligands and receptors. Brain Behav Evol 68: 181–190 - PubMed
-
- Alvarez Y, Alonso MT, Vendrell V, Zelarayan LC, Chamero P, Theil T, Bösl MR, Kato S, Maconochie M, Riethmacher D, et al. 2003. Requirements for FGF3 and FGF10 during inner ear formation. Development 130: 6329–6338 - PubMed
-
- Alvarez-Medina R, Cayuso J, Okubo T, Takada S, Marti E 2008. Wnt canonical pathway restricts graded Shh/Gli patterning activity through the regulation of Gli3 expression. Development 135: 237–247 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
