The recognition domain of the methyl-specific endonuclease McrBC flips out 5-methylcytosine
- PMID: 22570415
- PMCID: PMC3424535
- DOI: 10.1093/nar/gks332
The recognition domain of the methyl-specific endonuclease McrBC flips out 5-methylcytosine
Abstract
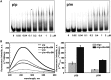
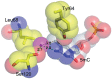
DNA cytosine methylation is a widespread epigenetic mark. Biological effects of DNA methylation are mediated by the proteins that preferentially bind to 5-methylcytosine (5mC) in different sequence contexts. Until now two different structural mechanisms have been established for 5mC recognition in eukaryotes; however, it is still unknown how discrimination of the 5mC modification is achieved in prokaryotes. Here we report the crystal structure of the N-terminal DNA-binding domain (McrB-N) of the methyl-specific endonuclease McrBC from Escherichia coli. The McrB-N protein shows a novel DNA-binding fold adapted for 5mC-recognition. In the McrB-N structure in complex with methylated DNA, the 5mC base is flipped out from the DNA duplex and positioned within a binding pocket. Base flipping elegantly explains why McrBC system restricts only T4-even phages impaired in glycosylation [Luria, S.E. and Human, M.L. (1952) A nonhereditary, host-induced variation of bacterial viruses. J. Bacteriol., 64, 557-569]: flipped out 5-hydroxymethylcytosine is accommodated in the binding pocket but there is no room for the glycosylated base. The mechanism for 5mC recognition employed by McrB-N is highly reminiscent of that for eukaryotic SRA domains, despite the differences in their protein folds.
Figures




References
-
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. - PubMed
-
- Tost J. DNA methylation: an introduction to the biology and the disease-associated changes of a promising biomarker. Mol. Biotechnol. 2010;44:71–81. - PubMed
-
- Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–424. - PubMed
-
- Ho KL, McNae IW, Schmiedeberg L, Klose RJ, Bird AP, Walkinshaw MD. MeCP2 binding to DNA depends upon hydration at methyl-CpG. Mol. Cell. 2008;29:525–531. - PubMed
-
- Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–821. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

