Active site opening and closure control translocation of multisubunit RNA polymerase
- PMID: 22570421
- PMCID: PMC3424550
- DOI: 10.1093/nar/gks383
Active site opening and closure control translocation of multisubunit RNA polymerase
Abstract
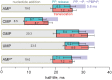
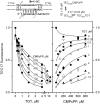
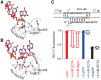
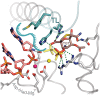
Multisubunit RNA polymerase (RNAP) is the central information-processing enzyme in all cellular life forms, yet its mechanism of translocation along the DNA molecule remains conjectural. Here, we report direct monitoring of bacterial RNAP translocation following the addition of a single nucleotide. Time-resolved measurements demonstrated that translocation is delayed relative to nucleotide incorporation and occurs shortly after or concurrently with pyrophosphate release. An investigation of translocation equilibrium suggested that the strength of interactions between RNA 3' nucleotide and nucleophilic and substrate sites determines the translocation state of transcription elongation complexes, whereas active site opening and closure modulate the affinity of the substrate site, thereby favoring the post- and pre-translocated states, respectively. The RNAP translocation mechanism is exploited by the antibiotic tagetitoxin, which mimics pyrophosphate and induces backward translocation by closing the active site.
Figures




Similar articles
-
Structural basis for transcription inhibition by tagetitoxin.Nat Struct Mol Biol. 2005 Dec;12(12):1086-93. doi: 10.1038/nsmb1015. Epub 2005 Nov 6. Nat Struct Mol Biol. 2005. PMID: 16273103 Free PMC article.
-
Tagetitoxin inhibits transcription by stabilizing pre-translocated state of the elongation complex.Nucleic Acids Res. 2013 Nov;41(20):9257-65. doi: 10.1093/nar/gkt708. Epub 2013 Aug 9. Nucleic Acids Res. 2013. PMID: 23935117 Free PMC article.
-
A small post-translocation energy bias aids nucleotide selection in T7 RNA polymerase transcription.Biophys J. 2012 Feb 8;102(3):532-41. doi: 10.1016/j.bpj.2011.12.028. Epub 2012 Feb 7. Biophys J. 2012. PMID: 22325276 Free PMC article.
-
Translocation by multi-subunit RNA polymerases.Biochim Biophys Acta. 2010 May-Jun;1799(5-6):389-401. doi: 10.1016/j.bbagrm.2010.01.007. Epub 2010 Jan 25. Biochim Biophys Acta. 2010. PMID: 20097318 Review.
-
Forks, pincers, and triggers: the tools for nucleotide incorporation and translocation in multi-subunit RNA polymerases.Curr Opin Struct Biol. 2009 Dec;19(6):708-14. doi: 10.1016/j.sbi.2009.10.008. Epub 2009 Nov 11. Curr Opin Struct Biol. 2009. PMID: 19913407 Free PMC article. Review.
Cited by
-
Rate-limiting Pyrophosphate Release by HIV Reverse Transcriptase Improves Fidelity.J Biol Chem. 2016 Dec 16;291(51):26554-26565. doi: 10.1074/jbc.M116.753152. Epub 2016 Oct 24. J Biol Chem. 2016. PMID: 27777304 Free PMC article.
-
The elemental mechanism of transcriptional pausing.Elife. 2019 Jan 8;8:e40981. doi: 10.7554/eLife.40981. Elife. 2019. PMID: 30618376 Free PMC article.
-
Interplay between the trigger loop and the F loop during RNA polymerase catalysis.Nucleic Acids Res. 2014 Jan;42(1):544-52. doi: 10.1093/nar/gkt877. Epub 2013 Oct 1. Nucleic Acids Res. 2014. PMID: 24089145 Free PMC article.
-
Millisecond dynamics of RNA polymerase II translocation at atomic resolution.Proc Natl Acad Sci U S A. 2014 May 27;111(21):7665-70. doi: 10.1073/pnas.1315751111. Epub 2014 Apr 21. Proc Natl Acad Sci U S A. 2014. PMID: 24753580 Free PMC article.
-
Regulation of Transcript Elongation.Annu Rev Microbiol. 2015;69:49-69. doi: 10.1146/annurev-micro-091014-104047. Epub 2015 Jun 24. Annu Rev Microbiol. 2015. PMID: 26132790 Free PMC article. Review.
References
-
- Steitz TA. A mechanism for all polymerases. Nature. 1998;391:231–232. - PubMed
-
- Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–168. - PubMed
-
- Chamberlin MJ. New models for the mechanism of transcription elongation and its regulation. Harvey Lect. 1992;88:1–21. - PubMed
-
- Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. - PubMed

