Active site opening and closure control translocation of multisubunit RNA polymerase
- PMID: 22570421
- PMCID: PMC3424550
- DOI: 10.1093/nar/gks383
Active site opening and closure control translocation of multisubunit RNA polymerase
Abstract
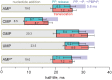
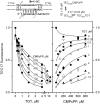
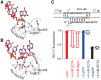
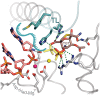
Multisubunit RNA polymerase (RNAP) is the central information-processing enzyme in all cellular life forms, yet its mechanism of translocation along the DNA molecule remains conjectural. Here, we report direct monitoring of bacterial RNAP translocation following the addition of a single nucleotide. Time-resolved measurements demonstrated that translocation is delayed relative to nucleotide incorporation and occurs shortly after or concurrently with pyrophosphate release. An investigation of translocation equilibrium suggested that the strength of interactions between RNA 3' nucleotide and nucleophilic and substrate sites determines the translocation state of transcription elongation complexes, whereas active site opening and closure modulate the affinity of the substrate site, thereby favoring the post- and pre-translocated states, respectively. The RNAP translocation mechanism is exploited by the antibiotic tagetitoxin, which mimics pyrophosphate and induces backward translocation by closing the active site.
Figures




References
-
- Steitz TA. A mechanism for all polymerases. Nature. 1998;391:231–232. - PubMed
-
- Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–168. - PubMed
-
- Chamberlin MJ. New models for the mechanism of transcription elongation and its regulation. Harvey Lect. 1992;88:1–21. - PubMed
-
- Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. - PubMed

