Structural and functional insight into how the Plasmodium falciparum VAR2CSA protein mediates binding to chondroitin sulfate A in placental malaria
- PMID: 22570492
- PMCID: PMC3390611
- DOI: 10.1074/jbc.M112.348839
Structural and functional insight into how the Plasmodium falciparum VAR2CSA protein mediates binding to chondroitin sulfate A in placental malaria
Abstract
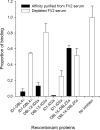
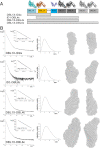
Malaria is a major global health problem. Pregnant women are susceptible to infection regardless of previously acquired immunity. Placental malaria is caused by parasites capable of sequestering in the placenta. This is mediated by VAR2CSA, a parasite antigen that interacts with chondroitin sulfate A (CSA). One vaccine strategy is to block this interaction with VAR2CSA-specific antibodies. It is a priority to define a small VAR2CSA fragment that can be used in an adhesion blocking vaccine. In this, the obvious approach is to define regions of VAR2CSA involved in receptor binding. It has been shown that full-length recombinant VAR2CSA binds specifically to CSA with nanomolar affinity, and that the CSA-binding site lies in the N-terminal part of the protein. In this study we define the minimal binding region by truncating VAR2CSA and analyzing CSA binding using biosensor technology. We show that the core CSA-binding site lies within the DBL2X domain and parts of the flanking interdomain regions. This is in contrast to the idea that single domains do not possess the structural requirements for specific CSA binding. Small-angle x-ray scattering measurements enabled modeling of VAR2CSA and showed that the CSA-binding DBL2X domain is situated in the center of the structure. Mutating classic sulfate-binding sites in VAR2CSA, along with testing dependence of ionic interactions, suggest that the CSA binding is not solely dependent on the sulfated CSA structure. Based on these novel PfEMP1 structure-function studies, we have constructed a small VAR2CSA antigen that has the capacity to induce highly adhesion-blocking antibodies.
Figures






References
-
- WHO (2010) World Malaria Report 2010, WHO, Geneva, Switzerland
-
- Fried M., Duffy P. E. (1996) Adherence of Plasmodium falciparum to chondroitin sulfate A in the human placenta. Science 272, 1502–1504 - PubMed
-
- Menendez C. (1995) Malaria during pregnancy. A priority area of malaria research and control. Parasitol. Today 11, 178–183 - PubMed
-
- McGregor I. A. (1987) Thoughts on malaria in pregnancy with consideration of some factors that influence remedial strategies. Parassitologia 29, 153–163 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

