Patterns of ancestry, signatures of natural selection, and genetic association with stature in Western African pygmies
- PMID: 22570615
- PMCID: PMC3343053
- DOI: 10.1371/journal.pgen.1002641
Patterns of ancestry, signatures of natural selection, and genetic association with stature in Western African pygmies
Abstract
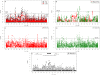
African Pygmy groups show a distinctive pattern of phenotypic variation, including short stature, which is thought to reflect past adaptation to a tropical environment. Here, we analyze Illumina 1M SNP array data in three Western Pygmy populations from Cameroon and three neighboring Bantu-speaking agricultural populations with whom they have admixed. We infer genome-wide ancestry, scan for signals of positive selection, and perform targeted genetic association with measured height variation. We identify multiple regions throughout the genome that may have played a role in adaptive evolution, many of which contain loci with roles in growth hormone, insulin, and insulin-like growth factor signaling pathways, as well as immunity and neuroendocrine signaling involved in reproduction and metabolism. The most striking results are found on chromosome 3, which harbors a cluster of selection and association signals between approximately 45 and 60 Mb. This region also includes the positional candidate genes DOCK3, which is known to be associated with height variation in Europeans, and CISH, a negative regulator of cytokine signaling known to inhibit growth hormone-stimulated STAT5 signaling. Finally, pathway analysis for genes near the strongest signals of association with height indicates enrichment for loci involved in insulin and insulin-like growth factor signaling.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Perry GH, Dominy NJ. Evolution of the human pygmy phenotype. Trends Ecol Evol. 2009;24:218–225. - PubMed
-
- Batini C, Lopes J, Behar DM, Calafell F, Jorde LB, et al. Insights into the demographic history of African Pygmies from complete mitochondrial genomes. Mol Biol Evol. 2011;28:1099–1110. - PubMed
-
- Destro-Bisol G, Coia V, Boschi I, Verginelli F, Caglia A, et al. The analysis of variation of mtDNA hypervariable region 1 suggests that Eastern and Western Pygmies diverged before the Bantu expansion. Am Nat. 2004;163:212–226. - PubMed
-
- Patin E, Laval G, Barreiro LB, Salas A, Semino O, et al. Inferring the demographic history of African farmers and pygmy hunter-gatherers using a multilocus resequencing data set. PLoS Genet. 2009;5:e1000448. doi: 10.1371/journal.pgen.1000448. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

