Sequence-specific targeting of dosage compensation in Drosophila favors an active chromatin context
- PMID: 22570616
- PMCID: PMC3343056
- DOI: 10.1371/journal.pgen.1002646
Sequence-specific targeting of dosage compensation in Drosophila favors an active chromatin context
Erratum in
- PLoS Genet. 2014 Jan;10(1). doi: 10.1371/annotation/39c76721-c8e1-4d42-ae6b-33063ce207d2 doi: 10.1371/annotation/39c76721-c8e1-4d42-ae6b-33063ce207d2
Abstract
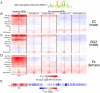
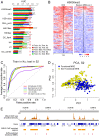
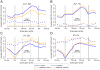
The Drosophila MSL complex mediates dosage compensation by increasing transcription of the single X chromosome in males approximately two-fold. This is accomplished through recognition of the X chromosome and subsequent acetylation of histone H4K16 on X-linked genes. Initial binding to the X is thought to occur at "entry sites" that contain a consensus sequence motif ("MSL recognition element" or MRE). However, this motif is only ∼2 fold enriched on X, and only a fraction of the motifs on X are initially targeted. Here we ask whether chromatin context could distinguish between utilized and non-utilized copies of the motif, by comparing their relative enrichment for histone modifications and chromosomal proteins mapped in the modENCODE project. Through a comparative analysis of the chromatin features in male S2 cells (which contain MSL complex) and female Kc cells (which lack the complex), we find that the presence of active chromatin modifications, together with an elevated local GC content in the surrounding sequences, has strong predictive value for functional MSL entry sites, independent of MSL binding. We tested these sites for function in Kc cells by RNAi knockdown of Sxl, resulting in induction of MSL complex. We show that ectopic MSL expression in Kc cells leads to H4K16 acetylation around these sites and a relative increase in X chromosome transcription. Collectively, our results support a model in which a pre-existing active chromatin environment, coincident with H3K36me3, contributes to MSL entry site selection. The consequences of MSL targeting of the male X chromosome include increase in nucleosome lability, enrichment for H4K16 acetylation and JIL-1 kinase, and depletion of linker histone H1 on active X-linked genes. Our analysis can serve as a model for identifying chromatin and local sequence features that may contribute to selection of functional protein binding sites in the genome.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

