Overexpression of the catalytically impaired Taspase1 T234V or Taspase1 D233A variants does not have a dominant negative effect in T(4;11) leukemia cells
- PMID: 22570686
- PMCID: PMC3343046
- DOI: 10.1371/journal.pone.0034142
Overexpression of the catalytically impaired Taspase1 T234V or Taspase1 D233A variants does not have a dominant negative effect in T(4;11) leukemia cells
Abstract
Background: The chromosomal translocation t(4;11)(q21;q23) is associated with high-risk acute lymphoblastic leukemia of infants. The resulting AF4•MLL oncoprotein becomes activated by Taspase1 hydrolysis and is considered to promote oncogenic transcriptional activation. Hence, Taspase1's proteolytic activity is a critical step in AF4•MLL pathophysiology. The Taspase1 proenzyme is autoproteolytically processed in its subunits and is assumed to assemble into an αββα-heterodimer, the active protease. Therefore, we investigated here whether overexpression of catalytically inactive Taspase1 variants are able to interfere with the proteolytic activity of the wild type enzyme in AF4•MLL model systems.
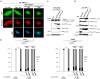
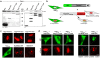
Methodology/findings: The consequences of overexpressing the catalytically dead Taspase1 mutant, Taspase1(T234V), or the highly attenuated variant, Taspase1(D233A), on Taspase1's processing of AF4•MLL and of other Taspase1 targets was analyzed in living cancer cells employing an optimized cell-based assay. Notably, even a nine-fold overexpression of the respective Taspase1 mutants neither inhibited Taspase1's cis- nor trans-cleavage activity in vivo. Likewise, enforced expression of the α- or β-subunits showed no trans-dominant effect against the ectopically or endogenously expressed enzyme. Notably, co-expression of the individual α- and β-subunits did not result in their assembly into an enzymatically active protease complex. Probing Taspase1 multimerization in living cells by a translocation-based protein interaction assay as well as by biochemical methods indicated that the inactive Taspase1 failed to assemble into stable heterocomplexes with the wild type enzyme.
Conclusions: Collectively, our results demonstrate that inefficient heterodimerization appears to be the mechanism by which inactive Taspase1 variants fail to inhibit wild type Taspase1's activity in trans. Our work favours strategies targeting Taspase1's catalytic activity rather than attempts to block the formation of active Taspase1 dimers to interfere with the pathobiological function of AF4•MLL.
Conflict of interest statement
Figures



References
-
- Meyer C, Kowarz E, Hofmann J, Renneville A, Zuna J, et al. New insights to the MLL recombinome of acute leukemias. Leukemia. 2009;23:1490–1499. - PubMed
-
- Montes R, Ayllon V, Gutierrez-Aranda I, Prat I, Hernandez-Lamas MC, et al. Enforced expression of MLL-AF4 fusion in cord blood CD34+ cells enhances the hematopoietic repopulating cell function and clonogenic potential but is not sufficient to initiate leukemia. Blood. 2011;117:4746–4758. - PubMed
-
- Bursen A, Schwabe K, Ruster B, Henschler R, Ruthardt M, et al. The AF4.MLL fusion protein is capable of inducing ALL in mice without requirement of MLL.AF4. Blood. 2010;115:3570–3579. - PubMed
-
- Yokoyama A, Kitabayashi I, Ayton PM, Cleary ML, Ohki M. Leukemia proto-oncoprotein MLL is proteolytically processed into 2 fragments with opposite transcriptional properties. Blood. 2002;100:3710–3718. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

