Colonial history and contemporary transmission shape the genetic diversity of hepatitis C virus genotype 2 in Amsterdam
- PMID: 22573865
- PMCID: PMC3416291
- DOI: 10.1128/JVI.06910-11
Colonial history and contemporary transmission shape the genetic diversity of hepatitis C virus genotype 2 in Amsterdam
Abstract
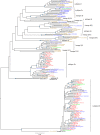
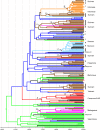
Evolutionary analysis of hepatitis C virus (HCV) genome sequences has provided insights into the epidemic history and transmission of this widespread human pathogen. Here we report an exceptionally diverse set of 178 HCV genotype 2 (HCV-2) isolates from 189 patients in Amsterdam, comprising 8 distinct HCV subtypes and 10 previously not recognized, unclassified lineages. By combining study subjects' demographic information with phylogeographic and molecular clock analyses, we demonstrate for the first time that the trans-Atlantic slave trade and colonial history were the driving forces behind the global dissemination of HCV-2. We detect multiple HCV-2 movements from present-day Ghana/Benin to the Caribbean during the peak years of the slave trade (1700 to 1850) and extensive transfer of HCV-2 among the Netherlands and its former colonies Indonesia and Surinam over the last 150 years. The latter coincides with the bidirectional migration of Javanese workers between Indonesia and Surinam and subsequent immigration to the Netherlands. In addition, our study sheds light on contemporary trends in HCV transmission within the Netherlands. We observe multiple lineages of the epidemic subtypes 2a, 2b, and 2c (together 67% of HCV-2 infections in Amsterdam), which cluster according to their suspected routes of transmission, specifically, injecting drug use (IDU) and contaminated blood/blood products. Understanding the epidemiological processes that generated the global pattern of HCV diversity seen today is critical for exposing associations between populations, risk factors, and specific HCV subtypes and might help HCV screening and prevention campaigns to minimize the future burden of HCV-related liver disease.
Figures


References
-
- Andriulli A, et al. 2008. Meta-analysis: the outcome of anti-viral therapy in HCV genotype 2 and genotype 3 infected patients with chronic hepatitis. Aliment. Pharmacol. Ther. 28:397–404 - PubMed
-
- Brucato N, et al. 2010. The imprint of the Slave Trade in an African American population: mitochondrial DNA, Y chromosome and HTLV-1 analysis in the Noir Marron of French Guiana. BMC Evol. Biol. 10:314 doi:10.1186/1471-2148-10-314 - DOI - PMC - PubMed
-
- Cantaloube J-F, et al. 2008. Molecular characterisation of genotype 2 and 4 hepatitis C virus isolates in French blood donors. J. Med. Virol. 80:1732–1739 - PubMed
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

