p53 Activation following Rift Valley fever virus infection contributes to cell death and viral production
- PMID: 22574148
- PMCID: PMC3344861
- DOI: 10.1371/journal.pone.0036327
p53 Activation following Rift Valley fever virus infection contributes to cell death and viral production
Abstract
Rift Valley fever virus (RVFV) is an emerging viral zoonosis that is responsible for devastating outbreaks among livestock and is capable of causing potentially fatal disease in humans. Studies have shown that upon infection, certain viruses have the capability of utilizing particular cellular signaling pathways to propagate viral infection. Activation of p53 is important for the DNA damage signaling cascade, initiation of apoptosis, cell cycle arrest and transcriptional regulation of multiple genes. The current study focuses on the role of p53 signaling in RVFV infection and viral replication. These results show an up-regulation of p53 phosphorylation at several serine sites after RVFV MP-12 infection that is highly dependent on the viral protein NSs. qRT-PCR data showed a transcriptional up-regulation of several p53 targeted genes involved in cell cycle and apoptosis regulation following RVFV infection. Cell viability assays demonstrate that loss of p53 results in less RVFV induced cell death. Furthermore, decreased viral titers in p53 null cells indicate that RVFV utilizes p53 to enhance viral production. Collectively, these experiments indicate that the p53 signaling pathway is utilized during RVFV infection to induce cell death and increase viral production.
Conflict of interest statement
Figures

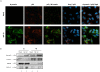



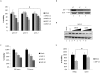
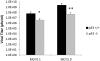
References
-
- Bird BH, Ksiazek TG, Nichol ST, Maclachlan NJ. Rift Valley fever virus. J Am Vet Med Assoc. 2009;234:883–893. - PubMed
-
- Daubney R, Hudson JR, Garnham PC. Enzootic hepatitis or rift valley fever. An undescribed virus disease of sheep cattle and man from east africa. The Journal of Pathology and Bacteriology. 1931;34:545–579.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

