Comparative SILAC proteomic analysis of Trypanosoma brucei bloodstream and procyclic lifecycle stages
- PMID: 22574199
- PMCID: PMC3344917
- DOI: 10.1371/journal.pone.0036619
Comparative SILAC proteomic analysis of Trypanosoma brucei bloodstream and procyclic lifecycle stages
Abstract
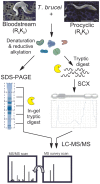
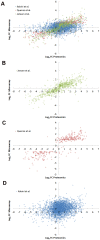
The protozoan parasite Trypanosoma brucei has a complex digenetic lifecycle between a mammalian host and an insect vector, and adaption of its proteome between lifecycle stages is essential to its survival and virulence. We have optimized a procedure for growing Trypanosoma brucei procyclic form cells in conditions suitable for stable isotope labeling by amino acids in culture (SILAC) and report a comparative proteomic analysis of cultured procyclic form and bloodstream form T. brucei cells. In total we were able to identify 3959 proteins and quantify SILAC ratios for 3553 proteins with a false discovery rate of 0.01. A large number of proteins (10.6%) are differentially regulated by more the 5-fold between lifecycle stages, including those involved in the parasite surface coat, and in mitochondrial and glycosomal energy metabolism. Our proteomic data is broadly in agreement with transcriptomic studies, but with significantly larger fold changes observed at the protein level than at the mRNA level.
Conflict of interest statement
Figures

References
-
- Wirtz E, Leal S, Ochatt C, Cross GAM. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol. 1999;99:89–101. - PubMed
-
- Carrington M. Parasites; 2009. How African trypanosomes evade mammalian defences. pp. 8–11.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

