Chloroplast RH3 DEAD box RNA helicases in maize and Arabidopsis function in splicing of specific group II introns and affect chloroplast ribosome biogenesis
- PMID: 22576849
- PMCID: PMC3387720
- DOI: 10.1104/pp.112.197525
Chloroplast RH3 DEAD box RNA helicases in maize and Arabidopsis function in splicing of specific group II introns and affect chloroplast ribosome biogenesis
Abstract
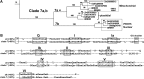
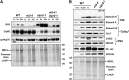
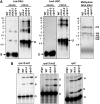
Chloroplasts in angiosperms contain at least seven nucleus-encoded members of the DEAD box RNA helicase family. Phylogenetic analysis shows that five of these plastid members (RH22, -39, -47, -50, and -58) form a single clade and that RH3 forms a clade with two mitochondrial RH proteins (PMH1 and -2) functioning in intron splicing. The function of chloroplast RH3 in maize (Zea mays; ZmRH3) and Arabidopsis (Arabidopsis thaliana; AtRH3) was determined. ZmRH3 and AtRH3 are both under strong developmental control, and ZmRH3 abundance sharply peaked in the sink-source transition zone of developing maize leaves, coincident with the plastid biogenesis machinery. ZmRH3 coimmunoprecipitated with a specific set of plastid RNAs, including several group II introns, as well as pre23S and 23S ribosomal RNA (rRNA), but not 16S rRNA. Furthermore, ZmRH3 associated with 50S preribosome particles as well as nucleoids. AtRH3 null mutants are embryo lethal, whereas a weak allele (rh3-4) results in pale-green seedlings with defects in splicing of several group II introns and rRNA maturation as well as reduced levels of assembled ribosomes. These results provide strong evidence that RH3 functions in the splicing of group II introns and possibly also contributes to the assembly of the 50S ribosomal particle. Previously, we observed 5- to 10-fold up-regulation of AtRH3 in plastid Caseinolytic protease mutants. The results shown here indicate that AtRH3 up-regulation was not a direct consequence of reduced proteolysis but constituted a compensatory response at both RH3 transcript and protein levels to impaired chloroplast biogenesis; this response demonstrates that cross talk between the chloroplast and the nucleus is used to regulate RH3 levels.
Figures







References
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

