Identification of novel inhibitors of M. tuberculosis growth using whole cell based high-throughput screening
- PMID: 22577943
- PMCID: PMC3560293
- DOI: 10.1021/cb300151m
Identification of novel inhibitors of M. tuberculosis growth using whole cell based high-throughput screening
Abstract
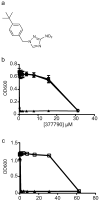
Despite the urgent need for new antitubercular drugs, few are on the horizon. To combat the problem of emerging drug resistance, structurally unique chemical entities that inhibit new targets will be required. Here we describe our investigations using whole cell screening of a diverse collection of small molecules as a methodology for identifying novel inhibitors that target new pathways for Mycobacterium tuberculosis drug discovery. We find that conducting primary screens using model mycobacterial species may limit the potential for identifying new inhibitors with efficacy against M. tuberculosis. In addition, we confirm the importance of developing in vitro assay conditions that are reflective of in vivo biology for maximizing the proportion of hits from whole cell screening that are likely to have activity in vivo. Finally, we describe the identification and characterization of two novel inhibitors that target steps in M. tuberculosis cell wall biosynthesis. The first is a novel benzimidazole that targets mycobacterial membrane protein large 3 (MmpL3), a proposed transporter for cell wall mycolic acids. The second is a nitro-triazole that inhibits decaprenylphosphoryl-β-D-ribose 2'-epimerase (DprE1), an epimerase required for cell wall biosynthesis. These proteins are both among the small number of new targets that have been identified by forward chemical genetics using resistance generation coupled with genome sequencing. This suggests that methodologies currently employed for screening and target identification may lead to a bias in target discovery and that alternative methods should be explored.
Figures




References
-
- Ma Z, Lienhardt C, McIlleron H, Nunn AJ, Wang X. Global tuberculosis drug development pipeline: the need and the reality. Lancet. 2010;375:2100–2109. - PubMed
-
- Andries K, Verhasselt P, Guillemont J, Göhlmann HWH, Neefs JM, Winkler H, Van Gestel J, Timmerman P, Zhu M, Lee E, Williams P, de Chaffoy D, Huitric E, Hoffner S, Cambau E, Truffot-Pernot C, Lounis N, Jarlier V. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science. 2005;307:223–227. - PubMed
-
- Sacchettini JC, Rubin EJ, Freundlich JS. Drugs versus bugs: in pursuit of the persistent predator Mycobacterium tuberculosis. Nat Rev Microbiol. 2008;6:41–52. - PubMed
-
- Keller TH, Pichota A, Yin Z. A practical view of ‘druggability’. Curr Opin Chem Biol. 2006;10:357–361. - PubMed
-
- Pethe K, Sequeira PC, Agarwalla S, Rhee K, Kuhen K, Phong WY, Patel V, Beer D, Walker JR, Duraiswamy J, Jiricek J, Keller TH, Chatterjee A, Tan MP, Ujjini M, Rao SPS, Camacho L, Bifani P, Mak PA, Ma I, Barnes SW, Chen Z, Plouffe D, Thayalan P, Ng SH, Au M, Lee BH, Tan BH, Ravindran S, Nanjundappa M, Lin X, Goh A, Lakshminarayana SB, Shoen C, Cynamon M, Kreiswirth B, Dartois V, Peters EC, Glynne R, Brenner S, Dick T. Nat Comms. Vol. 1. Nature Publishing Group; 2010. A chemical genetic screen in Mycobacterium tuberculosis identifies carbon-source-dependent growth inhibitors devoid of in vivo efficacy; pp. 1–8. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

