Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels
- PMID: 22579281
- PMCID: PMC6347373
- DOI: 10.1016/j.cell.2012.04.017
Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels
Abstract
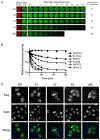
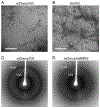
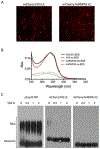
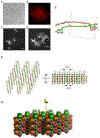
Eukaryotic cells contain assemblies of RNAs and proteins termed RNA granules. Many proteins within these bodies contain KH or RRM RNA-binding domains as well as low complexity (LC) sequences of unknown function. We discovered that exposure of cell or tissue lysates to a biotinylated isoxazole (b-isox) chemical precipitated hundreds of RNA-binding proteins with significant overlap to the constituents of RNA granules. The LC sequences within these proteins are both necessary and sufficient for b-isox-mediated aggregation, and these domains can undergo a concentration-dependent phase transition to a hydrogel-like state in the absence of the chemical. X-ray diffraction and EM studies revealed the hydrogels to be composed of uniformly polymerized amyloid-like fibers. Unlike pathogenic fibers, the LC sequence-based polymers described here are dynamic and accommodate heterotypic polymerization. These observations offer a framework for understanding the function of LC sequences as well as an organizing principle for cellular structures that are not membrane bound.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures




Comment in
-
Protein aggregation: The secret recipe for RNA granules.Nat Rev Mol Cell Biol. 2012 May 30;13(7):405. doi: 10.1038/nrm3372. Nat Rev Mol Cell Biol. 2012. PMID: 22644452 No abstract available.
-
Secrets of RNA granules.Nat Methods. 2012 Jul;9(7):639. doi: 10.1038/nmeth.2090. Nat Methods. 2012. PMID: 22930828 No abstract available.
References
-
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, and Hyman AA (2009). Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

