O-Glycosylation of snails
- PMID: 22581130
- PMCID: PMC3372779
- DOI: 10.1007/s10719-012-9391-4
O-Glycosylation of snails
Abstract
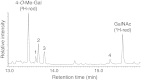
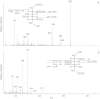
The glycosylation abilities of snails deserve attention, because snail species serve as intermediate hosts in the developmental cycles of some human and cattle parasites. In analogy to many other host-pathogen relations, the glycosylation of snail proteins may likewise contribute to these host-parasite interactions. Here we present an overview on the O-glycan structures of 8 different snails (land and water snails, with or without shell): Arion lusitanicus, Achatina fulica, Biomphalaria glabrata, Cepaea hortensis, Clea helena, Helix pomatia, Limax maximus and Planorbarius corneus. The O-glycans were released from the purified snail proteins by β-elimination. Further analysis was carried out by liquid chromatography coupled to electrospray ionization mass spectrometry and - for the main structures - by gas chromatography/mass spectrometry. Snail O-glycans are built from the four monosaccharide constituents: N-acetylgalactosamine, galactose, mannose and fucose. An additional modification is a methylation of the hexoses. The common trisaccharide core structure was determined in Arion lusitanicus to be N-acetylgalactosamine linked to the protein elongated by two 4-O-methylated galactose residues. Further elongations by methylated and unmethylated galactose and mannose residues and/or fucose are present. The typical snail O-glycan structures are different to those so far described. Similar to snail N-glycan structures they display methylated hexose residues.
Figures




References
-
- Patsos G, Corfield A. The sugar code. In: Gabius HJ, editor. pp. 111–137. Weinheim: Wiley-VCH; 2009.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

