Natural transformation of Gallibacterium anatis
- PMID: 22582057
- PMCID: PMC3416388
- DOI: 10.1128/AEM.00412-12
Natural transformation of Gallibacterium anatis
Abstract
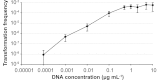
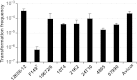
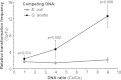
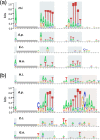
Gallibacterium anatis is a pathogen of poultry. Very little is known about its genetics and pathogenesis. To enable the study of gene function in G. anatis, we have established methods for transformation and targeted mutagenesis. The genus Gallibacterium belongs to the Pasteurellaceae, a group with several naturally transformable members, including Haemophilus influenzae. Bioinformatics analysis identified G. anatis homologs of the H. influenzae competence genes, and natural competence was induced in G. anatis by the procedure established for H. influenzae: transfer from rich medium to the starvation medium M-IV. This procedure gave reproducibly high transformation frequencies with G. anatis chromosomal DNA and with linearized plasmid DNA carrying G. anatis sequences. Both DNA types integrated into the G. anatis chromosome by homologous recombination. Targeted mutagenesis gave transformation frequencies of >2 × 10(-4) transformants CFU(-1). Transformation was also efficient with circular plasmid containing no G. anatis DNA; this resulted in the establishment of a self-replicating plasmid. Nine diverse G. anatis strains were found to be naturally transformable by this procedure, suggesting that natural competence is common and the M-IV transformation procedure widely applicable for this species. The G. anatis genome is only slightly enriched for the uptake signal sequences identified in other pasteurellaceaen genomes, but G. anatis did preferentially take up its own DNA over that of Escherichia coli. Transformation by electroporation was not effective for chromosomal integration but could be used to introduce self-replicating plasmids. The findings described here provide important tools for the genetic manipulation of G. anatis.
Figures

References
-
- Bonaventura MP, Lee EK, DeSalle R, Planet PJ. 2010. A whole-genome phylogeny of the family Pasteurellaceae. Mol. Phylogenet. Evol. 54: 950– 956 - PubMed
-
- Bossé JT, Nash JHE, Kroll JS, Langford PR. 2004. Harnessing natural transformation in Actinobacillus pleuropneumoniae: a simple method for allelic replacements. FEMS Microbiol. Lett. 233: 277– 281 - PubMed
-
- Bossé JT, et al. 2009. Natural competence in strains of Actinobacillus pleuropneumoniae. FEMS Microbiol. Lett. 298: 124– 130 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

