Exploration of multivariate analysis in microbial coding sequence modeling
- PMID: 22583558
- PMCID: PMC3473301
- DOI: 10.1186/1471-2105-13-97
Exploration of multivariate analysis in microbial coding sequence modeling
Abstract
Background: Gene finding is a complicated procedure that encapsulates algorithms for coding sequence modeling, identification of promoter regions, issues concerning overlapping genes and more. In the present study we focus on coding sequence modeling algorithms; that is, algorithms for identification and prediction of the actual coding sequences from genomic DNA. In this respect, we promote a novel multivariate method known as Canonical Powered Partial Least Squares (CPPLS) as an alternative to the commonly used Interpolated Markov model (IMM). Comparisons between the methods were performed on DNA, codon and protein sequences with highly conserved genes taken from several species with different genomic properties.
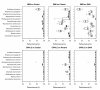
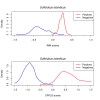
Results: The multivariate CPPLS approach classified coding sequence substantially better than the commonly used IMM on the same set of sequences. We also found that the use of CPPLS with codon representation gave significantly better classification results than both IMM with protein (p < 0.001) and with DNA (p < 0.001). Further, although the mean performance was similar, the variation of CPPLS performance on codon representation was significantly smaller than for IMM (p < 0.001).
Conclusions: The performance of coding sequence modeling can be substantially improved by using an algorithm based on the multivariate CPPLS method applied to codon or DNA frequencies.
Figures


Similar articles
-
Using hidden Markov models and observed evolution to annotate viral genomes.Bioinformatics. 2006 Jun 1;22(11):1308-16. doi: 10.1093/bioinformatics/btl092. Epub 2006 Apr 13. Bioinformatics. 2006. PMID: 16613911
-
Detecting overlapping coding sequences with pairwise alignments.Bioinformatics. 2005 Feb 1;21(3):282-92. doi: 10.1093/bioinformatics/bti007. Epub 2004 Sep 3. Bioinformatics. 2005. PMID: 15347574
-
Finding prokaryotic genes by the 'frame-by-frame' algorithm: targeting gene starts and overlapping genes.Bioinformatics. 1999 Nov;15(11):874-86. doi: 10.1093/bioinformatics/15.11.874. Bioinformatics. 1999. PMID: 10743554
-
[Analysis, identification and correction of some errors of model refseqs appeared in NCBI Human Gene Database by in silico cloning and experimental verification of novel human genes].Yi Chuan Xue Bao. 2004 May;31(5):431-43. Yi Chuan Xue Bao. 2004. PMID: 15478601 Chinese.
-
The computational detection of functional nucleotide sequence motifs in the coding regions of organisms.Exp Biol Med (Maywood). 2008 Jun;233(6):665-73. doi: 10.3181/0704-MR-97. Epub 2008 Apr 11. Exp Biol Med (Maywood). 2008. PMID: 18408149 Review.
Cited by
-
Comparative metabolomics of muscle interstitium fluid in human trapezius myalgia: an in vivo microdialysis study.Eur J Appl Physiol. 2013 Dec;113(12):2977-89. doi: 10.1007/s00421-013-2716-6. Eur J Appl Physiol. 2013. PMID: 24078209 Free PMC article.
-
Effect of chemotherapy on urinary volatile biomarkers for lung cancer by HS-SPME-GC-MS and chemometrics.Thorac Cancer. 2023 Dec;14(36):3522-3529. doi: 10.1111/1759-7714.15154. Epub 2023 Nov 9. Thorac Cancer. 2023. PMID: 37945317 Free PMC article.
-
Comparing K-mer based methods for improved classification of 16S sequences.BMC Bioinformatics. 2015 Jul 1;16:205. doi: 10.1186/s12859-015-0647-4. BMC Bioinformatics. 2015. PMID: 26130333 Free PMC article.
-
A systematic search for discriminating sites in the 16S ribosomal RNA gene.Microb Inform Exp. 2014 Jan 27;4(1):2. doi: 10.1186/2042-5783-4-2. Microb Inform Exp. 2014. PMID: 24467869 Free PMC article.
-
Feasibility of using volatile urine fingerprints for the differentiation of sexually transmitted infections.Appl Microbiol Biotechnol. 2023 Oct;107(20):6363-6376. doi: 10.1007/s00253-023-12711-0. Epub 2023 Aug 24. Appl Microbiol Biotechnol. 2023. PMID: 37615721 Free PMC article.
References
-
- Do J, Choi D. Computational approaches to gene prediction. J Microbiol Seoul. 2006;44(2):137. - PubMed
-
- Angelova M, Kalajdziski S, Kocarev L. Computational Methods for Gene Finding in Prokaryotes. Web Proceedings, ISSN. 2010;1:11–20.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

