Mouse strain specific gene expression differences for illumina microarray expression profiling in embryos
- PMID: 22583621
- PMCID: PMC3497855
- DOI: 10.1186/1756-0500-5-232
Mouse strain specific gene expression differences for illumina microarray expression profiling in embryos
Abstract
Background: In the field of mouse genetics the advent of technologies like microarray based expression profiling dramatically increased data availability and sensitivity, yet these advanced methods are often vulnerable to the unavoidable heterogeneity of in vivo material and might therefore reflect differentially expressed genes between mouse strains of no relevance to a targeted experiment. The aim of this study was not to elaborate on the usefulness of microarray analysis in general, but to expand our knowledge regarding this potential "background noise" for the widely used Illumina microarray platform surpassing existing data which focused primarily on the adult sensory and nervous system, by analyzing patterns of gene expression at different embryonic stages using wild type strains and modern transgenic models of often non-isogenic backgrounds.
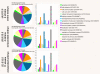
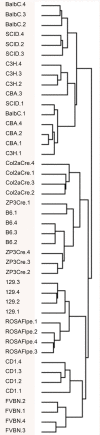
Results: Wild type embryos of 11 mouse strains commonly used in transgenic and molecular genetic studies at three developmental time points were subjected to Illumina microarray expression profiling in a strain-by-strain comparison. Our data robustly reflects known gene expression patterns during mid-gestation development. Decreasing diversity of the input tissue and/or increasing strain diversity raised the sensitivity of the array towards the genetic background. Consistent strain sensitivity of some probes was attributed to genetic polymorphisms or probe design related artifacts.
Conclusion: Our study provides an extensive reference list of gene expression profiling background noise of value to anyone in the field of developmental biology and transgenic research performing microarray expression profiling with the widely used Illumina microarray platform. Probes identified as strain specific background noise further allow for microarray expression profiling on its own to be a valuable tool for establishing genealogies of mouse inbred strains.
Figures


References
-
- Green EL. Biology of the laboratory mouse. New York: Dover Publication; 1966.
-
- Niswander L. Interplay between the molecular signals that control vertebrate limb development. Int J Dev Biol. 2002;46:877–881. - PubMed
-
- Zervas M, Blaess S, Joyner AL. Classical embryological studies and modern genetic analysis of midbrain and cerebellum development. Curr Top Dev Biol. 2005;69:101–138. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

