Development and application of a 6.5 million feature Affymetrix Genechip® for massively parallel discovery of single position polymorphisms in lettuce (Lactuca spp.)
- PMID: 22583801
- PMCID: PMC3490809
- DOI: 10.1186/1471-2164-13-185
Development and application of a 6.5 million feature Affymetrix Genechip® for massively parallel discovery of single position polymorphisms in lettuce (Lactuca spp.)
Abstract
Background: High-resolution genetic maps are needed in many crops to help characterize the genetic diversity that determines agriculturally important traits. Hybridization to microarrays to detect single feature polymorphisms is a powerful technique for marker discovery and genotyping because of its highly parallel nature. However, microarrays designed for gene expression analysis rarely provide sufficient gene coverage for optimal detection of nucleotide polymorphisms, which limits utility in species with low rates of polymorphism such as lettuce (Lactuca sativa).
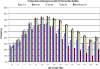
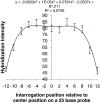
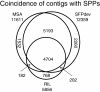
Results: We developed a 6.5 million feature Affymetrix GeneChip® for efficient polymorphism discovery and genotyping, as well as for analysis of gene expression in lettuce. Probes on the microarray were designed from 26,809 unigenes from cultivated lettuce and an additional 8,819 unigenes from four related species (L. serriola, L. saligna, L. virosa and L. perennis). Where possible, probes were tiled with a 2 bp stagger, alternating on each DNA strand; providing an average of 187 probes covering approximately 600 bp for each of over 35,000 unigenes; resulting in up to 13 fold redundancy in coverage per nucleotide. We developed protocols for hybridization of genomic DNA to the GeneChip® and refined custom algorithms that utilized coverage from multiple, high quality probes to detect single position polymorphisms in 2 bp sliding windows across each unigene. This allowed us to detect greater than 18,000 polymorphisms between the parental lines of our core mapping population, as well as numerous polymorphisms between cultivated lettuce and wild species in the lettuce genepool. Using marker data from our diversity panel comprised of 52 accessions from the five species listed above, we were able to separate accessions by species using both phylogenetic and principal component analyses. Additionally, we estimated the diversity between different types of cultivated lettuce and distinguished morphological types.
Conclusion: By hybridizing genomic DNA to a custom oligonucleotide array designed for maximum gene coverage, we were able to identify polymorphisms using two approaches for pair-wise comparisons, as well as a highly parallel method that compared all 52 genotypes simultaneously.
Figures





Comment in
-
An Ultra-High-Density, Transcript-Based, Genetic Map of Lettuce.G3 (Bethesda). 2013 Apr 9;3(4):617-631. doi: 10.1534/g3.112.004929. G3 (Bethesda). 2013. PMID: 23550116 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

