PCR-electrospray ionization mass spectrometry: the potential to change infectious disease diagnostics in clinical and public health laboratories
- PMID: 22584138
- PMCID: PMC7106027
- DOI: 10.1016/j.jmoldx.2012.02.005
PCR-electrospray ionization mass spectrometry: the potential to change infectious disease diagnostics in clinical and public health laboratories
Abstract
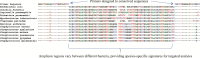
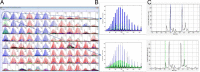
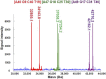
During the past 20 years, microbial detection methods that are genetically based, such as real-time PCR and peptide nucleic acid fluorescent hybridization, coexisted with traditional microbiological methods and were typically based on the identification of individual genetic targets. For these methods to be successful, a potential cause of infection must be suspected. More recently, multiplex PCR and multiplex RT-PCR were used to enable more broad-range testing based on panels of suspected pathogens. PCR-electrospray ionization mass spectrometry (PCR-ESI/MS) has emerged as a technology that is capable of identifying nearly all known human pathogens either from microbial isolates or directly from clinical specimens. Assay primers are strategically designed to target one or more of the broad pathogen categories: bacterial, mycobacterial, fungal, or viral. With broad-range amplification followed by detection of mixed amplicons, the method can identify genetic evidence of known and unknown pathogens. This unique approach supports a higher form of inquiry, asking the following question: What is the genetic evidence of known or unknown pathogens in the patient sample? This approach has advantages over traditional assays that commonly target the presence or absence of one or more pathogens with known genetic composition. This review considers the breadth of the published literature and explores the possibilities, advantages, and limitations for implementation of PCR-ESI/MS in diagnostic laboratories.
Copyright © 2012 American Society for Investigative Pathology and the Association for Molecular Pathology. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Hofstadler S.A., Sampath R., Blyn L.B., Eshoo M.W., Hall T.A., Jiang Y., Drader J.J., Hannis C., Sannes-Lwery K.A., Cummins L.L., Libby B., Knize D.J.D., Robbins D., Rudnik K., Desai A., Moradi E., Ecker D.J. TIGER: the universal biosensor. Int J Mass Spectrom. 2005;242:23–41.
-
- Ecker D.J., Sampath R., Willett P., Wyatt J.R., Samant V., Massire C., Hall T.A., Hari K., McNeil J.A., Buchen-Osmond C., Budowle B. The Microbial Rosetta Stone Database: a compilation of global and emerging infectious microorganisms and bioterrorist threat agents. BMC Microbiol. 2005;5:19. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

