PhiSpy: a novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies
- PMID: 22584627
- PMCID: PMC3439882
- DOI: 10.1093/nar/gks406
PhiSpy: a novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies
Abstract
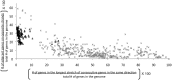
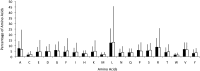
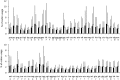
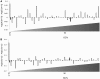
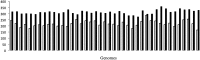
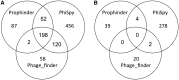
Prophages are phages in lysogeny that are integrated into, and replicated as part of, the host bacterial genome. These mobile elements can have tremendous impact on their bacterial hosts' genomes and phenotypes, which may lead to strain emergence and diversification, increased virulence or antibiotic resistance. However, finding prophages in microbial genomes remains a problem with no definitive solution. The majority of existing tools rely on detecting genomic regions enriched in protein-coding genes with known phage homologs, which hinders the de novo discovery of phage regions. In this study, a weighted phage detection algorithm, PhiSpy was developed based on seven distinctive characteristics of prophages, i.e. protein length, transcription strand directionality, customized AT and GC skew, the abundance of unique phage words, phage insertion points and the similarity of phage proteins. The first five characteristics are capable of identifying prophages without any sequence similarity with known phage genes. PhiSpy locates prophages by ranking genomic regions enriched in distinctive phage traits, which leads to the successful prediction of 94% of prophages in 50 complete bacterial genomes with a 6% false-negative rate and a 0.66% false-positive rate.
Figures

References
-
- Casjens S. Prophages and bacterial genomics: what have we learned so far? Mol. Microbiol. 2003;49:277–300. - PubMed
-
- Casjens S, Palmer N, van Vugt R, Huang WM, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson RJ, et al. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochaete Borrelia burgdorferi. Mol. Microbiol. 2000;35:490–516. - PubMed
-
- Mc Grath S, Van SD. Bacteriophage: Genetics and Molecular Biology. Norfolk, UK: Caister Academic Press; 2007.
-
- Aziz RK, Ismail S, Park HW, Kotb M. Post-proteomic identification of a novel phage-encoded streptodornase, Sda1, in invasive M1T1 Streptococcus pyogenes. Mol. Microbiol. 2004;54:184–197. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

