αE-catenin is an autoinhibited molecule that coactivates vinculin
- PMID: 22586082
- PMCID: PMC3365184
- DOI: 10.1073/pnas.1203906109
αE-catenin is an autoinhibited molecule that coactivates vinculin
Abstract
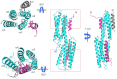
αE-catenin, an essential component of the adherens junction, interacts with the classical cadherin-β-catenin complex and with F-actin, but its precise role is unknown. αE-catenin also binds to the F-actin-binding protein vinculin, which also appears to be important in junction assembly. Vinculin and αE-catenin are homologs that contain a series of helical bundle domains, D1-D5. We mapped the vinculin-binding site to a sequence in D3a comprising the central two helices of a four-helix bundle. The crystal structure of this peptide motif bound to vinculin D1 shows that the two helices adopt a parallel, colinear arrangement suggesting that the αE-catenin D3a bundle must unfold in order to bind vinculin. We show that αE-catenin D3 binds strongly to vinculin, whereas larger fragments and full-length αE-catenin bind approximately 1,000-fold more weakly. Thus, intramolecular interactions within αE-catenin inhibit binding to vinculin. The actin-binding activity of vinculin is inhibited by an intramolecular interaction between the head (D1-D4) and the actin-binding D5 tail. In the absence of F-actin, there is no detectable binding of αE-catenin D3 to full-length vinculin; however, αE-catenin D3 promotes binding of vinculin to F-actin whereas full-length αE-catenin does not. These findings support the combinatorial or "coincidence" model of activation in which binding of high-affinity proteins to the vinculin head and tail is required to shift the conformational equilibrium of vinculin from a closed, autoinhibited state to an open, stable F-actin-binding state. The data also imply that αE-catenin must be activated in order to bind to vinculin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

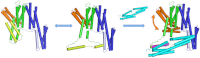

References
-
- Pokutta S, Weis WI. Structure and mechanism of cadherins and catenins in cell-cell contacts. Annu Rev Cell Dev Biol. 2007;23:237–261. - PubMed
-
- Dawes-Hoang RE, et al. Folded gastrulation, cell shape change and the control of myosin localization. Development. 2005;132:4165–4178. - PubMed
-
- Farge E. Mechanotransduction in development. Curr Top Dev Biol. 2011;95:243–265. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

