Superresolution imaging of HIV in infected cells with FlAsH-PALM
- PMID: 22586087
- PMCID: PMC3365178
- DOI: 10.1073/pnas.1013267109
Superresolution imaging of HIV in infected cells with FlAsH-PALM
Abstract
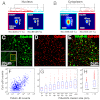
Imaging protein assemblies at molecular resolution without affecting biological function is a long-standing goal. The diffraction-limited resolution of conventional light microscopy (∼200-300 nm) has been overcome by recent superresolution (SR) methods including techniques based on accurate localization of molecules exhibiting stochastic fluorescence; however, SR methods still suffer important restrictions inherent to the protein labeling strategies. Antibody labels are encumbered by variable specificity, limited commercial availability and affinity, and are mostly restricted to fixed cells. Fluorescent protein fusions, though compatible with live cell imaging, substantially increase protein size and can interfere with their biological activity. We demonstrate SR imaging of proteins tagged with small tetracysteine motifs and the fluorescein arsenical helix binder (FlAsH-PALM). We applied FlAsH-PALM to image the integrase enzyme (IN) of HIV in fixed and living cells under experimental conditions that fully preserved HIV infectivity. The obtained resolution (∼30 nm) allowed us to characterize the distribution of IN within virions and intracellular complexes and to distinguish different HIV structural populations based on their morphology. We could thus discriminate ∼100 nm long mature conical cores from immature Gag shells and observe that in infected cells cytoplasmic (but not nuclear) IN complexes display a morphology similar to the conical capsid. Together with the presence of capsid proteins, our data suggest that cytoplasmic IN is largely present in intact capsids and that these can be found deep within the cytoplasm. FlAsH-PALM opens the door to in vivo SR studies of microbial complexes within host cells and may help achieve truly molecular resolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Knipe DM, Howley PM. Fundamental Virology. Philadelphia: Lippincott Williams and Wilkins; 2001.
-
- Hell SW. Microscopy and its focal switch. Nat Methods. 2009;6:24–32. - PubMed
-
- Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. - PubMed
-
- Fölling J, et al. Fluorescence nanoscopy by ground-state depletion and single-molecule return. Nat Methods. 2008;5:943–945. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

