Mass spectral molecular networking of living microbial colonies
- PMID: 22586093
- PMCID: PMC3387089
- DOI: 10.1073/pnas.1203689109
Mass spectral molecular networking of living microbial colonies
Abstract
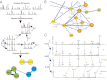
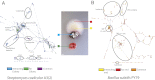
Integrating the governing chemistry with the genomics and phenotypes of microbial colonies has been a "holy grail" in microbiology. This work describes a highly sensitive, broadly applicable, and cost-effective approach that allows metabolic profiling of live microbial colonies directly from a Petri dish without any sample preparation. Nanospray desorption electrospray ionization mass spectrometry (MS), combined with alignment of MS data and molecular networking, enabled monitoring of metabolite production from live microbial colonies from diverse bacterial genera, including Bacillus subtilis, Streptomyces coelicolor, Mycobacterium smegmatis, and Pseudomonas aeruginosa. This work demonstrates that, by using these tools to visualize small molecular changes within bacterial interactions, insights can be gained into bacterial developmental processes as a result of the improved organization of MS/MS data. To validate this experimental platform, metabolic profiling was performed on Pseudomonas sp. SH-C52, which protects sugar beet plants from infections by specific soil-borne fungi [R. Mendes et al. (2011) Science 332:1097-1100]. The antifungal effect of strain SH-C52 was attributed to thanamycin, a predicted lipopeptide encoded by a nonribosomal peptide synthetase gene cluster. Our technology, in combination with our recently developed peptidogenomics strategy, enabled the detection and partial characterization of thanamycin and showed that it is a monochlorinated lipopeptide that belongs to the syringomycin family of antifungal agents. In conclusion, the platform presented here provides a significant advancement in our ability to understand the spatiotemporal dynamics of metabolite production in live microbial colonies and communities.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Comment in
-
A massively spectacular view of the chemical lives of microbes.Proc Natl Acad Sci U S A. 2012 Jun 26;109(26):10128-9. doi: 10.1073/pnas.1207725109. Epub 2012 Jun 18. Proc Natl Acad Sci U S A. 2012. PMID: 22711837 Free PMC article. No abstract available.
References
-
- Davies J. How to discover new antibiotics: harvesting the parvome. Curr Opin Chem Biol. 2011;15:5–10. - PubMed
-
- Davies J, Ryan KS. Introducing the parvome: Bioactive compounds in the microbial world. ACS Chem Biol. 2011;7:252–259. - PubMed
-
- Ratcliff WC, Denison RF. Microbiology. Alternative actions for antibiotics. Science. 2011;332:547–548. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

