Comparative genomics of rhizobia nodulating soybean suggests extensive recruitment of lineage-specific genes in adaptations
- PMID: 22586130
- PMCID: PMC3365164
- DOI: 10.1073/pnas.1120436109
Comparative genomics of rhizobia nodulating soybean suggests extensive recruitment of lineage-specific genes in adaptations
Abstract
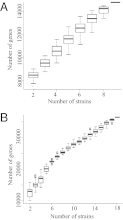
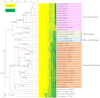
The rhizobium-legume symbiosis has been widely studied as the model of mutualistic evolution and the essential component of sustainable agriculture. Extensive genetic and recent genomic studies have led to the hypothesis that many distinct strategies, regardless of rhizobial phylogeny, contributed to the varied rhizobium-legume symbiosis. We sequenced 26 genomes of Sinorhizobium and Bradyrhizobium nodulating soybean to test this hypothesis. The Bradyrhizobium core genome is disproportionally enriched in lipid and secondary metabolism, whereas several gene clusters known to be involved in osmoprotection and adaptation to alkaline pH are specific to the Sinorhizobium core genome. These features are consistent with biogeographic patterns of these bacteria. Surprisingly, no genes are specifically shared by these soybean microsymbionts compared with other legume microsymbionts. On the other hand, phyletic patterns of 561 known symbiosis genes of rhizobia reflected the species phylogeny of these soybean microsymbionts and other rhizobia. Similar analyses with 887 known functional genes or the whole pan genome of rhizobia revealed that only the phyletic distribution of functional genes was consistent with the species tree of rhizobia. Further evolutionary genetics revealed that recombination dominated the evolution of core genome. Taken together, our results suggested that faithfully vertical genes were rare compared with those with history of recombination including lateral gene transfer, although rhizobial adaptations to symbiotic interactions and other environmental conditions extensively recruited lineage-specific shell genes under direct or indirect control through the speciation process.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Masson-Boivin C, Giraud E, Perret X, Batut J. Establishing nitrogen-fixing symbiosis with legumes: How many rhizobium recipes? Trends Microbiol. 2009;17:458–466. - PubMed
-
- Giraud E, et al. Legumes symbioses: Absence of Nod genes in photosynthetic bradyrhizobia. Science. 2007;316:1307–1312. - PubMed
-
- Mao C, Qiu J, Wang C, Charles TC, Sobral BW. NodMutDB: A database for genes and mutants involved in symbiosis. Bioinformatics. 2005;21:2927–2929. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

