NF-E2-related factor 1 (Nrf1) serves as a novel regulator of hepatic lipid metabolism through regulation of the Lipin1 and PGC-1β genes
- PMID: 22586274
- PMCID: PMC3416188
- DOI: 10.1128/MCB.06706-11
NF-E2-related factor 1 (Nrf1) serves as a novel regulator of hepatic lipid metabolism through regulation of the Lipin1 and PGC-1β genes
Abstract
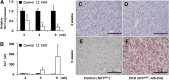
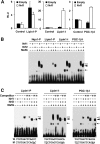
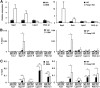
Hepatic lipid metabolism is under elaborate regulation, and perturbations in this regulatory process at the transcriptional level lead to pathological conditions. NF-E2-related factor 1 (Nrf1) is a member of the cap'n'collar (CNC) transcription factor family. Hepatocyte-specific Nrf1 gene conditional-knockout mice are known to develop hepatic steatosis, but it remains unclear how Nrf1 contributes to the lipid homeostasis. Therefore, in this study we examined the gene expression profiles of Nrf1-deficient mouse livers. A pathway analysis based on the profiling results revealed that the levels of expression of the genes related to lipid metabolism, amino acid metabolism, and mitochondrial respiratory function were decreased in Nrf1-deficient mouse livers, indicating the profound effects that the Nrf1 deficiency conferred to various metabolic pathways. We discovered that the Nrf1 deficiency leads to the reduced expression of the transcriptional coactivator genes Lipin1 and PGC-1β (for peroxisome proliferator-activated receptor γ coactivator 1β). Chromatin immunoprecipitation analyses showed that Nrf1 binds to the antioxidant response elements (AREs) in regulatory regions of the Lipin1 and PGC-1β genes and the binding of Nrf1 to the AREs activates reporter gene transcription. These results thus identified Nrf1 to be a novel regulator of the Lipin1 and PGC-1β genes, providing new insights into the Nrf1 function in hepatic lipid metabolism.
Figures





References
-
- Aoyama T, et al. 1998. Altered constitutive expression of fatty acid-metabolizing enzymes in mice lacking the peroxisome proliferator-activated receptor alpha (PPARalpha). J. Biol. Chem. 273:5678–5684 - PubMed
-
- Chowdhry S, et al. 2010. Loss of Nrf2 markedly exacerbates nonalcoholic steatohepatitis. Free Radic. Biol. Med. 48:357–371 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
