Tree-average distances on certain phylogenetic networks have their weights uniquely determined
- PMID: 22587565
- PMCID: PMC3395585
- DOI: 10.1186/1748-7188-7-13
Tree-average distances on certain phylogenetic networks have their weights uniquely determined
Abstract
A phylogenetic network N has vertices corresponding to species and arcs corresponding to direct genetic inheritance from the species at the tail to the species at the head. Measurements of DNA are often made on species in the leaf set, and one seeks to infer properties of the network, possibly including the graph itself. In the case of phylogenetic trees, distances between extant species are frequently used to infer the phylogenetic trees by methods such as neighbor-joining. This paper proposes a tree-average distance for networks more general than trees. The notion requires a weight on each arc measuring the genetic change along the arc. For each displayed tree the distance between two leaves is the sum of the weights along the path joining them. At a hybrid vertex, each character is inherited from one of its parents. We will assume that for each hybrid there is a probability that the inheritance of a character is from a specified parent. Assume that the inheritance events at different hybrids are independent. Then for each displayed tree there will be a probability that the inheritance of a given character follows the tree; this probability may be interpreted as the probability of the tree. The tree-average distance between the leaves is defined to be the expected value of their distance in the displayed trees. For a class of rooted networks that includes rooted trees, it is shown that the weights and the probabilities at each hybrid vertex can be calculated given the network and the tree-average distances between the leaves. Hence these weights and probabilities are uniquely determined. The hypotheses on the networks include that hybrid vertices have indegree exactly 2 and that vertices that are not leaves have a tree-child.
Keywords: digraph; distance; hybrid; metric; network; normal network; phylogeny; tree-child.
Figures
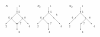

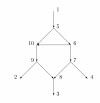
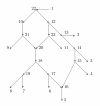
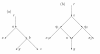

References
-
- Baroni M, Semple C, Steel M. A framework for representing reticulate evolution. Annals of Combinatorics. 2004;8:391–408.
-
- Nakhleh L, Warnow T, Linder CR. In: Proceedings of the Eighth Annual International Conference on Computational Molecular Biology (RECOMB '04 March 27-31, 2004. Bourne PE, Gusfield D, editor. San Diego, California), ACM, New York; 2004. Reconstructing reticulate evolution in species-theory and practice; pp. 337–346.
-
- Huson D, Rupp R, Scornavacca C. Phylogenetic Networks: Concepts, Algoriithms and Applications. Cambridge, Cambridge University Press; 2010.
LinkOut - more resources
Full Text Sources

