A practical approach to parameter estimation applied to model predicting heart rate regulation
- PMID: 22588357
- PMCID: PMC3526689
- DOI: 10.1007/s00285-012-0535-8
A practical approach to parameter estimation applied to model predicting heart rate regulation
Abstract
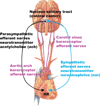
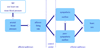
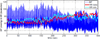
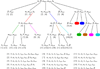
Mathematical models have long been used for prediction of dynamics in biological systems. Recently, several efforts have been made to render these models patient specific. One way to do so is to employ techniques to estimate parameters that enable model based prediction of observed quantities. Knowledge of variation in parameters within and between groups of subjects have potential to provide insight into biological function. Often it is not possible to estimate all parameters in a given model, in particular if the model is complex and the data is sparse. However, it may be possible to estimate a subset of model parameters reducing the complexity of the problem. In this study, we compare three methods that allow identification of parameter subsets that can be estimated given a model and a set of data. These methods will be used to estimate patient specific parameters in a model predicting baroreceptor feedback regulation of heart rate during head-up tilt. The three methods include: structured analysis of the correlation matrix, analysis via singular value decomposition followed by QR factorization, and identification of the subspace closest to the one spanned by eigenvectors of the model Hessian. Results showed that all three methods facilitate identification of a parameter subset. The "best" subset was obtained using the structured correlation method, though this method was also the most computationally intensive. Subsets obtained using the other two methods were easier to compute, but analysis revealed that the final subsets contained correlated parameters. In conclusion, to avoid lengthy computations, these three methods may be combined for efficient identification of parameter subsets.
Figures




References
-
- Anderson DA. Structural properties of compartmental models. Math Biosci. 1982;58:61–81.
-
- Astrom KJ, Cykhoff P. System identification-a survey. Automatica. 1971;7:123–162.
-
- Banks HT, Davidian M, Samuels JR, Sutton KL. In: An inverse problem statistical methodology summary, in Mathematical and statistical estimation approaches in Epidemiology. Chowell G, Hayman JM, Bettencourt LMA, Castillo-Chavez C, editors. New York, NY: Springer; 2009.
-
- Baker CTH, Paul CAH. Pitfalls in parameter estimation for delay differential equations. SIAM J Sci Comput. 1997;18:305–314.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

