Chimeras taking shape: potential functions of proteins encoded by chimeric RNA transcripts
- PMID: 22588898
- PMCID: PMC3396365
- DOI: 10.1101/gr.130062.111
Chimeras taking shape: potential functions of proteins encoded by chimeric RNA transcripts
Abstract
Chimeric RNAs comprise exons from two or more different genes and have the potential to encode novel proteins that alter cellular phenotypes. To date, numerous putative chimeric transcripts have been identified among the ESTs isolated from several organisms and using high throughput RNA sequencing. The few corresponding protein products that have been characterized mostly result from chromosomal translocations and are associated with cancer. Here, we systematically establish that some of the putative chimeric transcripts are genuinely expressed in human cells. Using high throughput RNA sequencing, mass spectrometry experimental data, and functional annotation, we studied 7424 putative human chimeric RNAs. We confirmed the expression of 175 chimeric RNAs in 16 human tissues, with an abundance varying from 0.06 to 17 RPKM (Reads Per Kilobase per Million mapped reads). We show that these chimeric RNAs are significantly more tissue-specific than non-chimeric transcripts. Moreover, we present evidence that chimeras tend to incorporate highly expressed genes. Despite the low expression level of most chimeric RNAs, we show that 12 novel chimeras are translated into proteins detectable in multiple shotgun mass spectrometry experiments. Furthermore, we confirm the expression of three novel chimeric proteins using targeted mass spectrometry. Finally, based on our functional annotation of exon organization and preserved domains, we discuss the potential features of chimeric proteins with illustrative examples and suggest that chimeras significantly exploit signal peptides and transmembrane domains, which can alter the cellular localization of cognate proteins. Taken together, these findings establish that some chimeric RNAs are translated into potentially functional proteins in humans.
Figures


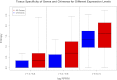
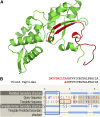
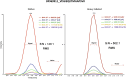
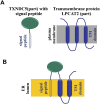
Comment in
-
Post transcriptional regulation: Chimeric protein production.Nat Rev Genet. 2012 Jun 7;13(7):450-1. doi: 10.1038/nrg3268. Nat Rev Genet. 2012. PMID: 22673160 No abstract available.
References
-
- Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, Spiegelman CH, Zimmerman LJ, Ham AJ, Keshishian H, et al. 2009. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol 27: 633–641 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
