Amino acid addition to Vibrio cholerae LPS establishes a link between surface remodeling in gram-positive and gram-negative bacteria
- PMID: 22589301
- PMCID: PMC3365186
- DOI: 10.1073/pnas.1201313109
Amino acid addition to Vibrio cholerae LPS establishes a link between surface remodeling in gram-positive and gram-negative bacteria
Abstract
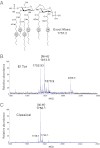
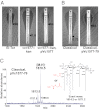
Historically, the O1 El Tor and classical biotypes of Vibrio cholerae have been differentiated by their resistance to the antimicrobial peptide polymyxin B. However, the molecular mechanisms associated with this phenotypic distinction have remained a mystery for 50 y. Both gram-negative and gram-positive bacteria modify their cell wall components with amine-containing substituents to reduce the net negative charge of the bacterial surface, thereby promoting cationic antimicrobial peptide resistance. In the present study, we demonstrate that V. cholerae modify the lipid A anchor of LPS with glycine and diglycine residues. This previously uncharacterized lipid A modification confers polymyxin resistance in V. cholerae El Tor, requiring three V. cholerae proteins: Vc1577 (AlmG), Vc1578 (AlmF), and Vc1579 (AlmE). Interestingly, the protein machinery required for glycine addition is reminiscent of the gram-positive system responsible for D-alanylation of teichoic acids. Such machinery was not thought to be used by gram-negative organisms. V. cholerae O1 El Tor mutants lacking genes involved in transferring glycine to LPS showed a 100-fold increase in sensitivity to polymyxin B. This work reveals a unique lipid A modification and demonstrates a charge-based remodeling strategy shared between gram-positive and gram-negative organisms.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Comment in
-
Bacterial physiology: Another brick in the wall.Nat Rev Microbiol. 2012 Jun 6;10(7):442. doi: 10.1038/nrmicro2817. Nat Rev Microbiol. 2012. PMID: 22669218 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

