MicroSCALE screening reveals genetic modifiers of therapeutic response in melanoma
- PMID: 22589389
- PMCID: PMC3498828
- DOI: 10.1126/scisignal.2002612
MicroSCALE screening reveals genetic modifiers of therapeutic response in melanoma
Abstract
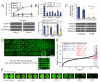
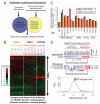
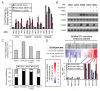
Cell microarrays are a promising tool for performing large-scale functional genomic screening in mammalian cells at reasonable cost, but owing to technical limitations they have been restricted for use with a narrow range of cell lines and short-term assays. Here, we describe MicroSCALE (Microarrays of Spatially Confined Adhesive Lentiviral Features), a cell microarray-based platform that enables application of this technology to a wide range of cell types and longer-term assays. We used MicroSCALE to uncover kinases that when overexpressed partially desensitized B-RAFV600E-mutant melanoma cells to inhibitors of the mitogen-activated protein kinase kinase kinase (MAPKKK) RAF, the MAPKKs MEK1 and 2 (MEK1/2, mitogen-activated protein kinase kinase 1 and 2), mTOR (mammalian target of rapamycin), or PI3K (phosphatidylinositol 3-kinase). These screens indicated that cells treated with inhibitors acting through common mechanisms were affected by a similar profile of overexpressed proteins. In contrast, screens involving inhibitors acting through distinct mechanisms yielded unique profiles, a finding that has potential relevance for small-molecule target identification and combination drugging studies. Further, by integrating large-scale functional screening results with cancer cell line gene expression and pharmacological sensitivity data, we validated the nuclear factor κB pathway as a potential mediator of resistance to MAPK pathway inhibitors. The MicroSCALE platform described here may enable new classes of large-scale, resource-efficient screens that were not previously feasible, including those involving combinations of cell lines, perturbations, and assay outputs or those involving limited numbers of cells and limited or expensive reagents.
Figures

References
-
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921. - PubMed
-
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, Carpenter AE, Foo SY, Stewart SA, Stockwell BR, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283. - PubMed
-
- Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, Frohling S, Chan EM, Sos ML, Michel K, Mermel C, Silver SJ, Weir BA, Reiling JH, Sheng Q, Gupta PB, Wadlow RC, Le H, Hoersch S, Wittner BS, Ramaswamy S, Livingston DM, Sabatini DM, Meyerson M, Thomas RK, Lander ES, Mesirov JP, Root DE, Gilliland DG, Jacks T, Hahn WC. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

