Grass microRNA gene paleohistory unveils new insights into gene dosage balance in subgenome partitioning after whole-genome duplication
- PMID: 22589464
- PMCID: PMC3442569
- DOI: 10.1105/tpc.112.095752
Grass microRNA gene paleohistory unveils new insights into gene dosage balance in subgenome partitioning after whole-genome duplication
Abstract
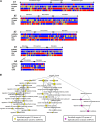
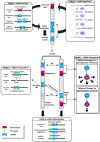
The recent availability of plant genome sequences, combined with a robust evolutionary scenario of the modern monocot and eudicot karyotypes from their diploid ancestors, offers an opportunity to gain insights into microRNA (miRNA) gene paleohistory in plants. Characterization and comparison of miRNAs and associated protein-coding targets in plants allowed us to unravel (1) contrasted genome conservation patterns of miRNAs in monocots and eudicots after whole-genome duplication (WGD), (2) an ancestral miRNA founder pool in the monocot genomes dating back to 100 million years ago, (3) miRNA subgenome dominance during the post-WGD diploidization process with selective miRNA deletion complemented with possible transposable element-mediated return flows, and (4) the miRNA/target interaction-directed differential loss/retention of miRNAs following the gene dosage balance rule. Together, our data suggest that overretained miRNAs in grass genomes may be implicated in connected gene regulations for stress responses, which is essential for plant adaptation and useful for crop variety innovation.
Figures




References
-
- Abrouk M., Murat F., Pont C., Messing J., Jackson S., Faraut T., Tannier E., Plomion C., Cooke R., Feuillet C., Salse J. (2010). Palaeogenomics of plants: Synteny-based modelling of extinct ancestors. Trends Plant Sci. 15: 479–487 - PubMed
-
- Allen E., Xie Z., Gustafson A.M., Sung G.H., Spatafora J.W., Carrington J.C. (2004). Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat. Genet. 36: 1282–1290 - PubMed
-
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410 - PubMed
-
- Ambros V. (2004). The functions of animal microRNAs. Nature 431: 350–355 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

