Systematic identification of functional plant modules through the integration of complementary data sources
- PMID: 22589469
- PMCID: PMC3387714
- DOI: 10.1104/pp.112.196725
Systematic identification of functional plant modules through the integration of complementary data sources
Erratum in
- Plant Physiol. 2012 aUG;159(4):1875
Abstract
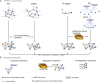
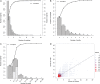
A major challenge is to unravel how genes interact and are regulated to exert specific biological functions. The integration of genome-wide functional genomics data, followed by the construction of gene networks, provides a powerful approach to identify functional gene modules. Large-scale expression data, functional gene annotations, experimental protein-protein interactions, and transcription factor-target interactions were integrated to delineate modules in Arabidopsis (Arabidopsis thaliana). The different experimental input data sets showed little overlap, demonstrating the advantage of combining multiple data types to study gene function and regulation. In the set of 1,563 modules covering 13,142 genes, most modules displayed strong coexpression, but functional and cis-regulatory coherence was less prevalent. Highly connected hub genes showed a significant enrichment toward embryo lethality and evidence for cross talk between different biological processes. Comparative analysis revealed that 58% of the modules showed conserved coexpression across multiple plants. Using module-based functional predictions, 5,562 genes were annotated, and an evaluation experiment disclosed that, based on 197 recently experimentally characterized genes, 38.1% of these functions could be inferred through the module context. Examples of confirmed genes of unknown function related to cell wall biogenesis, xylem and phloem pattern formation, cell cycle, hormone stimulus, and circadian rhythm highlight the potential to identify new gene functions. The module-based predictions offer new biological hypotheses for functionally unknown genes in Arabidopsis (1,701 genes) and six other plant species (43,621 genes). Furthermore, the inferred modules provide new insights into the conservation of coexpression and coregulation as well as a starting point for comparative functional annotation.
Figures




References
-
- Aoki K, Ogata Y, Shibata D. (2007) Approaches for extracting practical information from gene co-expression networks in plant biology. Plant Cell Physiol 48: 381–390 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

