Trace elements affect methanogenic activity and diversity in enrichments from subsurface coal bed produced water
- PMID: 22590465
- PMCID: PMC3349271
- DOI: 10.3389/fmicb.2012.00175
Trace elements affect methanogenic activity and diversity in enrichments from subsurface coal bed produced water
Abstract
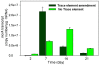
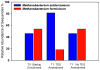
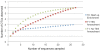
Microbial methane from coal beds accounts for a significant and growing percentage of natural gas worldwide. Our knowledge of physical and geochemical factors regulating methanogenesis is still in its infancy. We hypothesized that in these closed systems, trace elements (as micronutrients) are a limiting factor for methanogenic growth and activity. Trace elements are essential components of enzymes or cofactors of metabolic pathways associated with methanogenesis. This study examined the effects of eight trace elements (iron, nickel, cobalt, molybdenum, zinc, manganese, boron, and copper) on methane production, on mcrA transcript levels, and on methanogenic community structure in enrichment cultures obtained from coal bed methane (CBM) well produced water samples from the Powder River Basin, Wyoming. Methane production was shown to be limited both by a lack of additional trace elements as well as by the addition of an overly concentrated trace element mixture. Addition of trace elements at concentrations optimized for standard media enhanced methane production by 37%. After 7 days of incubation, the levels of mcrA transcripts in enrichment cultures with trace element amendment were much higher than in cultures without amendment. Transcript levels of mcrA correlated positively with elevated rates of methane production in supplemented enrichments (R(2) = 0.95). Metabolically active methanogens, identified by clone sequences of mcrA mRNA retrieved from enrichment cultures, were closely related to Methanobacterium subterraneum and Methanobacterium formicicum. Enrichment cultures were dominated by M. subterraneum and had slightly higher predicted methanogenic richness, but less diversity than enrichment cultures without amendments. These results suggest that varying concentrations of trace elements in produced water from different subsurface coal wells may cause changing levels of CBM production and alter the composition of the active methanogenic community.
Keywords: coal bed methane; enrichments; mcrA transcript; methanogens; trace elements.
Figures



References
-
- Basiliko N., Yavitt J. B. (2001). Influence of Ni, Co, Fe, and Na additions on methane production in Sphagnum-dominated Northern American peatlands. Biogeochemistry 52, 133–15310.1023/A:1006461803585 - DOI
-
- Becerra C. A., Lopez-Luna E., Ergas S. J., Nüsslein K. (2009). Microcosm-based study of the attenuation of an acid mine drainage-impacted site through biological sulfate and iron reduction, Geomicrobiol. J. 26, 9–2010.1080/01490450802599250 - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

