A genome-wide siRNA screen to identify modulators of insulin sensitivity and gluconeogenesis
- PMID: 22590537
- PMCID: PMC3348929
- DOI: 10.1371/journal.pone.0036384
A genome-wide siRNA screen to identify modulators of insulin sensitivity and gluconeogenesis
Abstract
Background: Hepatic insulin resistance impairs insulin's ability to suppress hepatic glucose production (HGP) and contributes to the development of type 2 diabetes (T2D). Although the interests to discover novel genes that modulate insulin sensitivity and HGP are high, it remains challenging to have a human cell based system to identify novel genes.
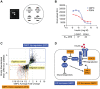
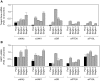
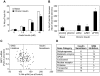
Methodology/principal findings: To identify genes that modulate hepatic insulin signaling and HGP, we generated a human cell line stably expressing beta-lactamase under the control of the human glucose-6-phosphatase (G6PC) promoter (AH-G6PC cells). Both beta-lactamase activity and endogenous G6PC mRNA were increased in AH-G6PC cells by a combination of dexamethasone and pCPT-cAMP, and reduced by insulin. A 4-gene High-Throughput-Genomics assay was developed to concomitantly measure G6PC and pyruvate-dehydrogenase-kinase-4 (PDK4) mRNA levels. Using this assay, we screened an siRNA library containing pooled siRNA targeting 6650 druggable genes and identified 614 hits that lowered G6PC expression without increasing PDK4 mRNA levels. Pathway analysis indicated that siRNA-mediated knockdown (KD) of genes known to positively or negatively affect insulin signaling increased or decreased G6PC mRNA expression, respectively, thus validating our screening platform. A subset of 270 primary screen hits was selected and 149 hits were confirmed by target gene KD by pooled siRNA and 7 single siRNA for each gene to reduce G6PC expression in 4-gene HTG assay. Subsequently, pooled siRNA KD of 113 genes decreased PEPCK and/or PGC1alpha mRNA expression thereby demonstrating their role in regulating key gluconeogenic genes in addition to G6PC. Last, KD of 61 of the above 113 genes potentiated insulin-stimulated Akt phosphorylation, suggesting that they suppress gluconeogenic gene by enhancing insulin signaling.
Conclusions/significance: These results support the proposition that the proteins encoded by the genes identified in our cell-based druggable genome siRNA screen hold the potential to serve as novel pharmacological targets for the treatment of T2D.
Conflict of interest statement
Figures



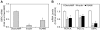

References
-
- Saltiel AR. New perspectives into the molecular pathogenesis and treatment of type 2 diabetes. Cell. 2001;104:517–529. - PubMed
-
- Yang R, Trevillyan JM. c-Jun N-terminal kinase pathways in diabetes. Int J Biochem Cell Biol. 2008;40:2702–2706. - PubMed
-
- Zimmet P, Alberti KG, Shaw J. Global and societal implications of the diabetes epidemic. Nature. 2001;414:782–787. - PubMed
-
- Mithieux G, Guignot L, Bordet JC, Wiernsperger N. Intrahepatic mechanisms underlying the effect of metformin in decreasing basal glucose production in rats fed a high-fat diet. Diabetes. 2002;51:139–143. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

