Pseudomonas syringae pv. actinidiae (PSA) isolates from recent bacterial canker of kiwifruit outbreaks belong to the same genetic lineage
- PMID: 22590555
- PMCID: PMC3348921
- DOI: 10.1371/journal.pone.0036518
Pseudomonas syringae pv. actinidiae (PSA) isolates from recent bacterial canker of kiwifruit outbreaks belong to the same genetic lineage
Abstract
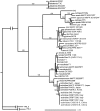
Intercontinental spread of emerging plant diseases is one of the most serious threats to world agriculture. One emerging disease is bacterial canker of kiwi fruit (Actinidia deliciosa and A. chinensis) caused by Pseudomonas syringae pv. actinidiae (PSA). The disease first occurred in China and Japan in the 1980s and in Korea and Italy in the 1990s. A more severe form of the disease broke out in Italy in 2008 and in additional countries in 2010 and 2011 threatening the viability of the global kiwi fruit industry. To start investigating the source and routes of international transmission of PSA, genomes of strains from China (the country of origin of the genus Actinidia), Japan, Korea, Italy and Portugal have been sequenced. Strains from China, Italy, and Portugal have been found to belong to the same clonal lineage with only 6 single nucleotide polymorphisms (SNPs) in 3,453,192 bp and one genomic island distinguishing the Chinese strains from the European strains. Not more than two SNPs distinguish each of the Italian and Portuguese strains from each other. The Japanese and Korean strains belong to a separate genetic lineage as previously reported. Analysis of additional European isolates and of New Zealand isolates exploiting genome-derived markers showed that these strains belong to the same lineage as the Italian and Chinese strains. Interestingly, the analyzed New Zealand strains are identical to European strains at the tested SNP loci but test positive for the genomic island present in the sequenced Chinese strains and negative for the genomic island present in the European strains. Results are interpreted in regard to the possible direction of movement of the pathogen between countries and suggest a possible Chinese origin of the European and New Zealand outbreaks.
Conflict of interest statement
Figures


References
-
- Dye DW, Bradbury JF, Goto M, Hayward AC, Lelliott RA, et al. International standards for naming pathovars of phytopathogenic bacteria and a list of pathovar names and pathotype strains. Rev Plant Pathol. 1980;142:153–158.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

