Chitin binding proteins act synergistically with chitinases in Serratia proteamaculans 568
- PMID: 22590591
- PMCID: PMC3348882
- DOI: 10.1371/journal.pone.0036714
Chitin binding proteins act synergistically with chitinases in Serratia proteamaculans 568
Abstract
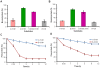
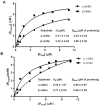
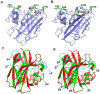
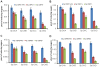
Genome sequence of Serratia proteamaculans 568 revealed the presence of three family 33 chitin binding proteins (CBPs). The three Sp CBPs (Sp CBP21, Sp CBP28 and Sp CBP50) were heterologously expressed and purified. Sp CBP21 and Sp CBP50 showed binding preference to β-chitin, while Sp CBP28 did not bind to chitin and cellulose substrates. Both Sp CBP21 and Sp CBP50 were synergistic with four chitinases from S. proteamaculans 568 (Sp ChiA, Sp ChiB, Sp ChiC and Sp ChiD) in degradation of α- and β-chitin, especially in the presence of external electron donor (reduced glutathione). Sp ChiD benefited most from Sp CBP21 or Sp CBP50 on α-chitin, while Sp ChiB and Sp ChiD had major advantage with these Sp CBPs on β-chitin. Dose responsive studies indicated that both the Sp CBPs exhibit synergism ≥ 0.2 µM. The addition of both Sp CBP21 and Sp CBP50 in different ratios to a synergistic mixture did not significantly increase the activity. Highly conserved polar residues, important in binding and activity of CBP21 from S. marcescens (Sm CBP21), were present in Sp CBP21 and Sp CBP50, while Sp CBP28 had only one such polar residue. The inability of Sp CBP28 to bind to the test substrates could be attributed to the absence of important polar residues.
Conflict of interest statement
Figures

References
-
- Rinaudo M. Chitin and chitosan: Properties and applications. Prog Polym Sci. 2006;31:603–632.
-
- Neeraja Ch, Anil K, Purushotham P, Suma K, Sarma PVSRN, et al. Biotechnological approaches to develop bacterial chitinases as a bioshield against fungal diseases of plants. Crit Rev Biotechnol. 2010c;30:231–241. - PubMed
-
- Zamil SS, Ahmad S, Choi MH, Yoon SC. Production of poly-N-acetylglucosamine by Staphylococcus saprophyticus BMSZ711: Characterization and production optimization. Bioresour Technol. 2010;101:7177–7180. - PubMed
-
- Vaaje-Kolstad G, Bunæs AC, Mathiesen G, Eijsink VGH. The chitinolytic system of Lactococcus lactis ssp. Lactis comprises a nonprocessive chitinase and a chitin-binding protein that promotes the degradation of α- and β-chitin. FEBS J. 2009;276:2402–2415. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

