Human peripheral blood antibodies with long HCDR3s are established primarily at original recombination using a limited subset of germline genes
- PMID: 22590602
- PMCID: PMC3348910
- DOI: 10.1371/journal.pone.0036750
Human peripheral blood antibodies with long HCDR3s are established primarily at original recombination using a limited subset of germline genes
Abstract
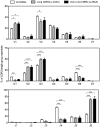
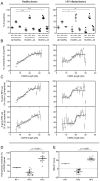
A number of antibodies that efficiently neutralize microbial targets contain long heavy chain complementarity determining region 3 (HCDR3) loops. For HIV, several of the most broad and potently neutralizing antibodies have exceptionally long HCDR3s. Two broad potently neutralizing HIV-specific antibodies, PG9 and PG16, exhibit secondary structure. Two other long HCDR3 antibodies, 2F5 and 4E10, protect against mucosal challenge with SHIV. Induction of such long HCDR3 antibodies may be critical to the design of an effective vaccine strategy for HIV and other pathogens, however it is unclear at present how to induce such antibodies. Here, we present genetic evidence that human peripheral blood antibodies containing long HCDR3s are not primarily generated by insertions introduced during the somatic hypermutation process. Instead, they are typically formed by processes occurring as part of the original recombination event. Thus, the response of B cells encoding antibodies with long HCDR3s results from selection of unusual clones from the naïve repertoire rather than through accumulation of insertions. These antibodies typically use a small subset of D and J gene segments that are particularly suited to encoding long HCDR3s, resulting in the incorporation of highly conserved genetic elements in the majority of antibody sequences encoding long HCDR3s.
Conflict of interest statement
Figures



References
-
- Stijlemans B, Conrath K, Cortez-Retamozo V, Van Xong H, Wyns L, et al. Efficient targeting of conserved cryptic epitopes of infectious agents by single domain antibodies. African trypanosomes as paradigm. J Biol Chem. 2004;279:1256–1261. doi: 10.1074/jbc.M307341200. - DOI - PubMed
-
- Henderson KA, Streltsov VA, Coley AM, Dolezal O, Hudson PJ, et al. Structure of an IgNAR-AMA1 complex: targeting a conserved hydrophobic cleft broadens malarial strain recognition. Structure. 2007;15:1452–1466. doi: 10.1016/j.str.2007.09.011. - DOI - PubMed
-
- Burton DR, Stanfield RL, Wilson IA. Antibody vs. HIV in a clash of evolutionary titans. Proc Natl Acad Sci USA. 2005;102:14943–14948. doi: 10.1073/pnas.0505126102. - DOI - PMC - PubMed
-
- Walker LM, Phogat SK, Chan-Hui P-Y, Wagner D, Phung P, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. - DOI - PMC - PubMed
-
- Pancera M, McLellan JS, Wu X, Zhu J, Changela A, et al. Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1. J Virol. 2010;84:8098–8110. doi: 10.1128/JVI.00966-10. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

