Structural basis for differential neutralization of ebolaviruses
- PMID: 22590681
- PMCID: PMC3347318
- DOI: 10.3390/v4040447
Structural basis for differential neutralization of ebolaviruses
Abstract
There are five antigenically distinct ebolaviruses that cause hemorrhagic fever in humans or non-human primates (Ebola virus, Sudan virus, Reston virus, Taï Forest virus, and Bundibugyo virus). The small handful of antibodies known to neutralize the ebolaviruses bind to the surface glycoprotein termed GP₁,₂. Curiously, some antibodies against them are known to neutralize in vitro but not protect in vivo, whereas other antibodies are known to protect animal models in vivo, but not neutralize in vitro. A detailed understanding of what constitutes a neutralizing and/or protective antibody response is critical for development of novel therapeutic strategies. Here, we show that paradoxically, a lower affinity antibody with restricted access to its epitope confers better neutralization than a higher affinity antibody against a similar epitope, suggesting that either subtle differences in epitope, or different characteristics of the GP₁,₂ molecules themselves, confer differential neutralization susceptibility. Here, we also report the crystal structure of trimeric, prefusion GP₁,₂ from the original 1976 Boniface variant of Sudan virus complexed with 16F6, the first antibody known to neutralize Sudan virus, and compare the structure to that of Sudan virus, variant Gulu. We discuss new structural details of the GP₁-GP₂ clamp, thermal motion of various regions in GP₁,₂ across the two viruses visualized, details of differential interaction of the crystallized neutralizing antibodies, and their relevance for virus neutralization.
Keywords: Ebola; Filovirus; Sudan virus; antibodies; ebolavirus; neutralization: glycoprotein; structure.
Figures


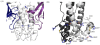




References
-
- Sanchez A., Khan A.S., Zaki S.R., Nabel G.J., Ksiazek T.G., Peters C.J. Filoviridae: Marburg and Ebola viruses. In: Knipe D.M., Howley P.M., editors. Fields Virology. Lippincott, Williams, and Wilkins; Philadelphia: 2001. pp. 1279–1304.
-
- Towner J.S., Sealy T.K., Khristova M.L., Albarino C.G., Conlan S., Reeder S.A., Quan P.L., Lipkin W.I., Downing R., Tappero J.W., Okware S., Lutwama J., Bakamutumaho B., Kayiwa J., Comer J.A., Rollin P.E., Ksiazek T.G., Nichol S.T. Newly discovered ebola virus associated with hemorrhagic fever outbreak in Uganda. PLoS Pathog. 2008;4:e1000212. - PMC - PubMed
-
- Sanchez A., Rollin P.E. Complete genome sequence of an Ebola virus (Sudan species) responsible for a 2000 outbreak of human disease in Uganda. Virus Res. 2005;113:16–25. - PubMed
-
- Maruyama T., Parren P.W., Sanchez A., Rensink I., Rodriguez L.L., Khan A.S., Peters C.J., Burton D.R. Recombinant human monoclonal antibodies to Ebola virus. J. Infect. Dis. 1999;179(Suppl 1):S235–S239. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- NS070899/NS/NINDS NIH HHS/United States
- AI081982/AI/NIAID NIH HHS/United States
- R01 AI081982/AI/NIAID NIH HHS/United States
- AI088027/AI/NIAID NIH HHS/United States
- AI068730/AI/NIAID NIH HHS/United States
- AI072106/AI/NIAID NIH HHS/United States
- R01 AI088027/AI/NIAID NIH HHS/United States
- GM020501/GM/NIGMS NIH HHS/United States
- U01 AI070530/AI/NIAID NIH HHS/United States
- R21 AI082437/AI/NIAID NIH HHS/United States
- R01 NS070899/NS/NINDS NIH HHS/United States
- GM093325/GM/NIGMS NIH HHS/United States
- R01 GM093325/GM/NIGMS NIH HHS/United States
- AI067927/AI/NIAID NIH HHS/United States
- R01 GM020501/GM/NIGMS NIH HHS/United States
- S10 RR029388/RR/NCRR NIH HHS/United States
- R01 AI067927/AI/NIAID NIH HHS/United States
- AI082437/AI/NIAID NIH HHS/United States
- F32 GM020501/GM/NIGMS NIH HHS/United States
- GM066170/GM/NIGMS NIH HHS/United States
- R56 AI088027/AI/NIAID NIH HHS/United States
- AI2008031/AI/NIAID NIH HHS/United States
- RR029388/RR/NCRR NIH HHS/United States
- T32 AI070117/AI/NIAID NIH HHS/United States
- AI070530/AI/NIAID NIH HHS/United States
- P01 AI068730/AI/NIAID NIH HHS/United States
- R01 AI072106/AI/NIAID NIH HHS/United States
- R01 GM066170/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

