Histone deacetylase (HDAC) inhibitors targeting HDAC3 and HDAC1 ameliorate polyglutamine-elicited phenotypes in model systems of Huntington's disease
- PMID: 22590724
- PMCID: PMC3528106
- DOI: 10.1016/j.nbd.2012.01.016
Histone deacetylase (HDAC) inhibitors targeting HDAC3 and HDAC1 ameliorate polyglutamine-elicited phenotypes in model systems of Huntington's disease
Abstract
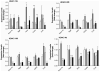
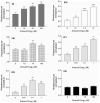
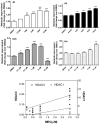
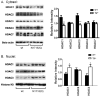
We have previously demonstrated amelioration of Huntington's disease (HD)-related phenotypes in R6/2 transgenic mice in response to treatment with the novel histone deacetylase (HDAC) inhibitor 4b. Here we have measured the selectivity profiles of 4b and related compounds against class I and class II HDACs and have tested their ability to restore altered expression of genes related to HD pathology in mice and to rescue disease effects in cell culture and Drosophila models of HD. R6/2 transgenic and wild-type (wt) mice received daily injections of HDAC inhibitors for 3 days followed by real-time PCR analysis to detect expression differences for 13 HD-related genes. We find that HDACi 4b and 136, two compounds showing high potency for inhibiting HDAC3 were most effective in reversing the expression of genes relevant to HD, including Ppp1r1b, which encodes DARPP-32, a marker for medium spiny striatal neurons. In contrast, compounds targeting HDAC1 were less effective at correcting gene expression abnormalities in R6/2 transgenic mice, but did cause significant increases in the expression of selected genes. An additional panel of 4b-related compounds was tested in a Drosophila model of HD and in STHdhQ111 striatal cells to further distinguish HDAC selectivity. Significant improvement in huntingtin-elicited Drosophila eye neurodegeneration in the fly was observed in response to treatment with compounds targeting human HDAC1 and/or HDAC3. In STHdhQ111 striatal cells, the ability of HDAC inhibitors to improve huntingtin-elicited metabolic deficits correlated with the potency at inhibiting HDAC1 and HDAC3, although the IC50 values for HDAC1 inhibition were typically 10-fold higher than for inhibition of HDAC3. Assessment of HDAC protein localization in brain tissue by Western blot analysis revealed accumulation of HDAC1 and HDAC3 in the nucleus of HD transgenic mice compared to wt mice, with a concurrent decrease in cytoplasmic localization, suggesting that these HDACs contribute to a repressive chromatin environment in HD. No differences were detected in the localization of HDAC2, HDAC4 or HDAC7. These results suggest that inhibition of HDACs 1 and 3 can relieve HD-like phenotypes in model systems and that HDAC inhibitors targeting these isotypes might show therapeutic benefit in human HD.
Figures
References
-
- Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–35. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous
