MetaMapp: mapping and visualizing metabolomic data by integrating information from biochemical pathways and chemical and mass spectral similarity
- PMID: 22591066
- PMCID: PMC3495401
- DOI: 10.1186/1471-2105-13-99
MetaMapp: mapping and visualizing metabolomic data by integrating information from biochemical pathways and chemical and mass spectral similarity
Abstract
Background: Exposure to environmental tobacco smoke (ETS) leads to higher rates of pulmonary diseases and infections in children. To study the biochemical changes that may precede lung diseases, metabolomic effects on fetal and maternal lungs and plasma from rats exposed to ETS were compared to filtered air control animals. Genome- reconstructed metabolic pathways may be used to map and interpret dysregulation in metabolic networks. However, mass spectrometry-based non-targeted metabolomics datasets often comprise many metabolites for which links to enzymatic reactions have not yet been reported. Hence, network visualizations that rely on current biochemical databases are incomplete and also fail to visualize novel, structurally unidentified metabolites.
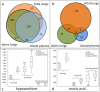
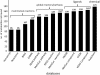
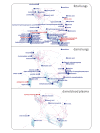
Results: We present a novel approach to integrate biochemical pathway and chemical relationships to map all detected metabolites in network graphs (MetaMapp) using KEGG reactant pair database, Tanimoto chemical and NIST mass spectral similarity scores. In fetal and maternal lungs, and in maternal blood plasma from pregnant rats exposed to environmental tobacco smoke (ETS), 459 unique metabolites comprising 179 structurally identified compounds were detected by gas chromatography time of flight mass spectrometry (GC-TOF MS) and BinBase data processing. MetaMapp graphs in Cytoscape showed much clearer metabolic modularity and complete content visualization compared to conventional biochemical mapping approaches. Cytoscape visualization of differential statistics results using these graphs showed that overall, fetal lung metabolism was more impaired than lungs and blood metabolism in dams. Fetuses from ETS-exposed dams expressed lower lipid and nucleotide levels and higher amounts of energy metabolism intermediates than control animals, indicating lower biosynthetic rates of metabolites for cell division, structural proteins and lipids that are critical for in lung development.
Conclusions: MetaMapp graphs efficiently visualizes mass spectrometry based metabolomics datasets as network graphs in Cytoscape, and highlights metabolic alterations that can be associated with higher rate of pulmonary diseases and infections in children prenatally exposed to ETS. The MetaMapp scripts can be accessed at http://metamapp.fiehnlab.ucdavis.edu.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

