DIAPH3 governs the cellular transition to the amoeboid tumour phenotype
- PMID: 22593025
- PMCID: PMC3494074
- DOI: 10.1002/emmm.201200242
DIAPH3 governs the cellular transition to the amoeboid tumour phenotype
Abstract
Therapies for most malignancies are generally ineffective once metastasis occurs. While tumour cells migrate through tissues using diverse strategies, the signalling networks controlling such behaviours in human tumours are poorly understood. Here we define a role for the Diaphanous-related formin-3 (DIAPH3) as a non-canonical regulator of metastasis that restrains conversion to amoeboid cell behaviour in multiple cancer types. The DIAPH3 locus is close to RB1, within a narrow consensus region of deletion on chromosome 13q in prostate, breast and hepatocellular carcinomas. DIAPH3 silencing in human carcinoma cells destabilized microtubules and induced defective endocytic trafficking, endosomal accumulation of EGFR, and hyperactivation of EGFR/MEK/ERK signalling. Silencing also evoked amoeboid properties, increased invasion and promoted metastasis in mice. In human tumours, DIAPH3 down-regulation was associated with aggressive or metastatic disease. DIAPH3-silenced cells were sensitive to MEK inhibition, but showed reduced sensitivity to EGFR inhibition. These findings have implications for understanding mechanisms of metastasis, and suggest that identifying patients with chromosomal deletions at DIAPH3 may have prognostic value.
Copyright © 2012 EMBO Molecular Medicine.
Figures

A. The DIAPH3 locus on chromosome 13q21.2 is located in a focal peak region of significant deletion (q ≤ 0.25) common in prostate, hepatocellular and breast carcinomas. Only three genes, PCDH17, TDRD3 and DIAPH3, are focally deleted in all cancer subtypes.
B-D. DIAPH3 mRNA levels in prostate (PCa, stage T2/T3), hepatocellular and invasive breast carcinomas compared to normal tissue.
E. Frequency of loss at DIAPH3 (grey line) in DTCs aspirated from bone marrow of patients with prostate-confined cancer (n = 48, blue) and advanced PCa (n = 11, red). Determined by array CGH and compared to copy number status in the primary tumours (n = 9, green). The chromosome band for each genomic position is shown; tick marks above indicate gene locations on chromosome 13.
F. DIAPH3 copy number status grouped by Gleason score and compared with metastases.
G. DIAPH3 copy number analysis in matched primary tumours and metastases (Met) reveals significant loss in metastatic disease.
H. DIAPH3 gene copy number in PCa samples grouped by Gleason Score in comparison to metastases (Met).
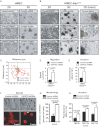
Morphology of DIAPH3-depleted HMEC, compared to control cells. Plastic (2D), basement membrane cultures (3D). Scale = 200 µm.
HRASV12 cells grown in 2D adopt a round, refractile appearance when DIAPH3 is silenced. Scale = 100 µm. HMEC-HRASV12 cells form cell aggregates with cord-like protrusions in 3D. DIAPH3 depletion causes junctional instability, with migration of single cells into the surrounding matrix (inset, right). Scale = 200 µm.
Representative trajectories of control (green) and DIAPH3-deficient (red) HMEC-HRASV12 cells.
Quantification of migration speed, determined from C.
DIAPH3-silenced HRASV12-HMEC display increased invasiveness.
Growth of DIAPH3-deficient and control DU145 cells on a thick layer of collagen I. Arrowheads mark cell surface blebbing (top). Visualization of the cytoskeleton with phalloidin by immunofluorescence (IF) shows prominent cortical actin in DIAPH3-deficient cells (arrowhead, bottom). Scale = 50 µm.
Increase in amoeboid morphology following expression of DIAPH3 siRNA.
Invasion of collagen I by DU145 cells, with FBS or EGF as chemo-attractants, is increased in DIAPH3-deficient cells (N ≥ 2 independent trials). See also Supporting Information Movies S1 and S2.
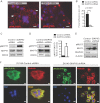
A. The distribution of focal adhesions in COS7 cells, detected by IF with a phospho-FAK(Y397) antibody, is reduced in DIAPH3-deficient cells. Scale = 20 µm.
B. Quantitation of focal adhesions in A.
C-E. Enhanced phosphorylation of MYPT1 (C,D) and MLC2 (E) in DIAPH3-deficient DU145 cells indicates high ROCK activity.
F. Distribution of active (phosphorylated) MLC2 and F-actin (phalloidin) in DU145 stable lines. Scale = 10 µm (N ≥ 2 independent trials).
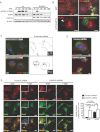
Enforced DIAPH3 accelerates EGFR turnover. COS7 cells expressing empty vector (Vo) or DIAPH3 were serum-depleted overnight, treated with EGF for the indicated times, and lysates blotted with the antibodies shown.
GFP-DIAPH3-positive COS7 cells show strongly reduced EGFR levels (lower panel, arrowhead) in comparison with GFP-transfected control cells (upper panel) or adjacent untransfected cells. Scale = 20 µm.
Left, Co-localization of Cy3-labeled EGFR with FITC-labelled Rab11 in DU145 cells expressing DIAPH3 siRNAs. Right, Cell peripheries are shown in grey, and areas of co-localization in black. Insets show predominance of EGFR at the plasma membrane (top) or in endosomes (bottom). Scale = 20 µm.
Left, Co-localization of Cy3-labeled EGFR with FITC-labelled Rab5 upon ligand binding (+EGF). DIAPH3 loss evokes an increase in EGFR internalization in the absence of ligand (−EGF), as shown by accumulation in endosomes (insets and arrowheads) and co-localization with Rab5. Scale = 20 µm. Right, Quantitation of EGFR-enriched endosomes.
Cy3-labeled EGFR, which is localized at the plasma membrane in DU145 cells expressing control siRNA, is internalized and co-localizes with the FITC-labelled early endosome marker EEA1 in DU145 cells expressing DIAPH3 siRNAs. Scale = 10 µm (N ≥ 2 independent trials).
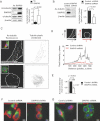
A. Increased acetylated tubulin (Ac-tubulin) in PC3 cells stably expressing DIAPH3.
B. Reduced Ac-tubulin levels in DU145 cells silenced for DIAPH3.
C. Left, Ac-tubulin IF showing MT fragmentation in DIAPH3-deficient DU145 cells grown in 3D. Right, MT topology was rendered by ImageJ. Insets, cells stained with cholera toxin B (CTxB, green). Scale = 10 µm.
D. Pixel intensity of C was quantified as a 2D contour plot, as a function of intensity in the x versus y planes (illustrated schematically, top).
E. The pixel intensity of Ac-tubulin was quantified in silenced or unsilenced DU145 cells. Cell peripheries were outlined as in C, tubulin intensity integrated within the enclosed area, and average intensities in each condition determined. Average intensity values determined from three independent trials (Student's t-test, p = 0.0033). N = total cell number.
F,G. DU145 cells expressing control or DIAPH3 shRNAs were stained with Ac-tubulin following staining with rhodamine-phalloidin (F) or FITC-CTxB (G), and imaged by fluorescence microscopy. Scale = 10 µm (N ≥ 2 independent trials).

EGFR is active in DIAPH3-silenced COS7 cells, as shown by increased tyrosine phosphorylation at activating sites and decreased phosphorylation of the T669 inhibitory site.
Enhanced pMEK1/2 in DIAPH3-depleted DU145 cells.
Targeting of DIAPH3 with four independent siRNAs in COS7 leads to >90% depletion, which inversely correlates with pERK1/2 levels.
DIAPH3 depletion in serum-depleted cells enhances pERK1/2 levels.
Acute, sustained phosphorylation of ERK1/2 in response to EGF in DIAPH3-depleted versus control cells (two-way ANOVA, p = 0.0068; N ≥ 2 independent trials).
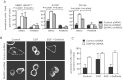
A. Amoeboid features induced by DIAPH3 silencing in human HMEC-HRASV12, A375P melanoma and DU145 cells can be reverted by PD98059 (50 µM). N ≥ 2 independent trials.
B,C. Amoeboid blebbing induced by EGF (10 nM) in control cells is sensitive to the EGFR inhibitor Gefitinib (2 µM), while induction by DIAPH3 silencing is less sensitive to both treatments, (C) Representative micrographs, (B) See also Supporting Information Movie S3.
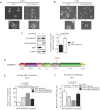
A,B. Micrographs of PC3 (A) and U87 (B) cells expressing GFP or GFP-DIAPH3. Arrows indicate prevalence of amoeboid cells in PC3- or U87-GFP (left panels), and of mesenchymal (lamellopodia-enriched) cells in PC3- or U87-GFP-DIAPH3 (right panels).
C. N-cadherin is significantly upregulated in cells expressing GFP-DIAPH3.
D. Domain structure of DIAPH3 and location of the S624 phosphosite within the FH1 domain.
E. Quantitation of stress fiber formation in HeLa cells expressing GFP or DIAPH3 mutants and stained with phalloidin.
F. Quantitation of amoeboid blebbing in U87 cells expressing GFP or DIAPH3 mutants and stained with CTxB (N ≥ 2 independent trials).
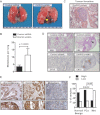
DIAPH3-silenced DU145 cells injected into the tail vein of nude mice produced large superficial pulmonary metastases (arrowhead).
Quantification of lung metastases in mice injected with control or DIAPH3-silenced DU145 cells (Mann–Whitney U test). N = 10 mice/condition.
Representative thromboembolus from lung sections from mice injected with DIAPH3-silenced DU145 cells.
Representative lung sections of mice injected with cells expressing control or DIAPH3 shRNAs, stained with H&E or an antibody to human CK18. Note presence of micrometastases in sections from DIAPH3 shRNA mice (arrowhead; N = 2 independent trials).
DIAPH3 IHC staining of a human tissue microarray (TMA) containing cores with benign human prostate epithelium (Normal/Benign), prostate tumour tissue (Carcinoma) and tissue from metastatic lesions (Metastasis). High-power magnifications are shown (bottom). Scale = 200 µm.
DIAPH3 expression is significantly decreased in metastases (Fisher's exact test, p = 0.018/0.007).
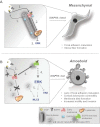
When DIAPH3 is functional: (1) EGFR is internalized via endocytosis upon activation; (2) EGFR traffics in endosomes along microtubules to lysosomes or is recycled back to the plasma membrane; (3) ERK activity is attenuated following EGFR down-regulation.
When DIAPH3 is lost or inactivated: (1) EGFR is internalized. (2) MT are destabilized, preventing both transport of EGFR from early endosomes to lysosomes, and receptor recycling. (3) Active EGFR accumulates in endosomes. (4) ERK activity is sustained. (5) Deregulation of proteins promoting amoeboid characteristics (e.g. MLC2, FAK) evokes cortical actomyosin contraction, focal adhesion turnover, and membrane blebbing. Broken arrows/lines indicate the presence of signalling intermediates.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

