Tension-dependent nucleosome remodeling at the pericentromere in yeast
- PMID: 22593210
- PMCID: PMC3386219
- DOI: 10.1091/mbc.E11-07-0651
Tension-dependent nucleosome remodeling at the pericentromere in yeast
Abstract
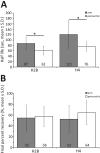
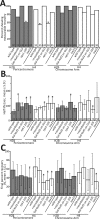
Nucleosome positioning is important for the structural integrity of chromosomes. During metaphase the mitotic spindle exerts physical force on pericentromeric chromatin. The cell must adjust the pericentromeric chromatin to accommodate the changing tension resulting from microtubule dynamics to maintain a stable metaphase spindle. Here we examine the effects of spindle-based tension on nucleosome dynamics by measuring the histone turnover of the chromosome arm and the pericentromere during metaphase in the budding yeast Saccharomyces cerevisiae. We find that both histones H2B and H4 exhibit greater turnover in the pericentromere during metaphase. Loss of spindle-based tension by treatment with the microtubule-depolymerizing drug nocodazole or compromising kinetochore function results in reduced histone turnover in the pericentromere. Pericentromeric histone dynamics are influenced by the chromatin-remodeling activities of STH1/NPS1 and ISW2. Sth1p is the ATPase component of the Remodels the Structure of Chromatin (RSC) complex, and Isw2p is an ATP-dependent DNA translocase member of the Imitation Switch (ISWI) subfamily of chromatin-remodeling factors. The balance between displacement and insertion of pericentromeric histones provides a mechanism to accommodate spindle-based tension while maintaining proper chromatin packaging during mitosis.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

