Centromere remodeling in Hoolock leuconedys (Hylobatidae) by a new transposable element unique to the gibbons
- PMID: 22593550
- PMCID: PMC3606032
- DOI: 10.1093/gbe/evs048
Centromere remodeling in Hoolock leuconedys (Hylobatidae) by a new transposable element unique to the gibbons
Abstract
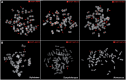
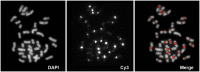
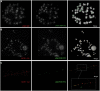
Gibbons (Hylobatidae) shared a common ancestor with the other hominoids only 15-18 million years ago. Nevertheless, gibbons show very distinctive features that include heavily rearranged chromosomes. Previous observations indicate that this phenomenon may be linked to the attenuated epigenetic repression of transposable elements (TEs) in gibbon species. Here we describe the massive expansion of a repeat in almost all the centromeres of the eastern hoolock gibbon (Hoolock leuconedys). We discovered that this repeat is a new composite TE originating from the combination of portions of three other elements (L1ME5, AluSz6, and SVA_A) and thus named it LAVA. We determined that this repeat is found in all the gibbons but does not occur in other hominoids. Detailed investigation of 46 different LAVA elements revealed that the majority of them have target site duplications (TSDs) and a poly-A tail, suggesting that they have been retrotransposing in the gibbon genome. Although we did not find a direct correlation between the emergence of LAVA elements and human-gibbon synteny breakpoints, this new composite transposable element is another mark of the great plasticity of the gibbon genome. Moreover, the centromeric expansion of LAVA insertions in the hoolock closely resembles the massive centromeric expansion of the KERV-1 retroelement reported for wallaby (marsupial) interspecific hybrids. The similarity between the two phenomena is consistent with the hypothesis that evolution of the gibbons is characterized by defects in epigenetic repression of TEs, perhaps triggered by interspecific hybridization.
Figures

References
-
- Arnold ML, Meyer A. Natural hybridization in primates: one evolutionary mechanism. Zoology. 2006;109:261–276. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

