Dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during embryogenesis
- PMID: 22593555
- PMCID: PMC3460198
- DOI: 10.1101/gr.134833.111
Dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during embryogenesis
Abstract
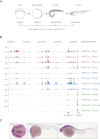
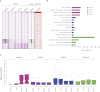
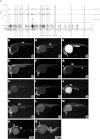
The generation of distinctive cell types that form different tissues and organs requires precise, temporal and spatial control of gene expression. This depends on specific cis-regulatory elements distributed in the noncoding DNA surrounding their target genes. Studies performed on mammalian embryonic stem cells and Drosophila embryos suggest that active enhancers form part of a defined chromatin landscape marked by histone H3 lysine 4 mono-methylation (H3K4me1) and histone H3 lysine 27 acetylation (H3K27ac). Nevertheless, little is known about the dynamics and the potential roles of these marks during vertebrate embryogenesis. Here, we provide genomic maps of H3K4me1/me3 and H3K27ac at four developmental time-points of zebrafish embryogenesis and analyze embryonic enhancer activity. We find that (1) changes in H3K27ac enrichment at enhancers accompany the shift from pluripotency to tissue-specific gene expression, (2) in early embryos, the peaks of H3K27ac enrichment are bound by pluripotent factors such as Nanog, and (3) the degree of evolutionary conservation is higher for enhancers that become marked by H3K27ac at the end of gastrulation, suggesting their implication in the establishment of the most conserved (phylotypic) transcriptome that is known to occur later at the pharyngula stage.
Figures


References
-
- Bauer S, Grossmann S, Vingron M, Robinson PN 2008. Ontologizer 2.0—a multifunctional tool for GO term enrichment analysis and data exploration. Bioinformatics 24: 1650–1651 - PubMed
-
- Bessa J, Tena JJ, de la Calle-Mustienes E, Fernandez-Minan A, Naranjo S, Fernandez A, Montoliu L, Akalin A, Lenhard B, Casares F, et al. 2009. Zebrafish enhancer detection (ZED) vector: A new tool to facilitate transgenesis and the functional analysis of cis-regulatory regions in zebrafish. Dev Dyn 238: 2409–2417 - PubMed
-
- Blader P, Lam CS, Rastegar S, Scardigli R, Nicod JC, Simplicio N, Plessy C, Fischer N, Schuurmans C, Guillemot F, et al. 2004. Conserved and acquired features of neurogenin1 regulation. Development 131: 5627–5637 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
